import numpy as np
import simpy
from sim_tools.distributions import ExponentialInitialisation bias
Learning objectives:
- Understand initialisation bias and know options for handling it.
- Add a warm-up period to a model.
Pre-reading:
This page continues on from: Entity processing.
Entity generation → Entity processing → Initialisation bias
Required packages:
These should be available from environment setup in the “Test yourself” section of Environments.
library(dplyr)
library(simmer)Initialisation bias
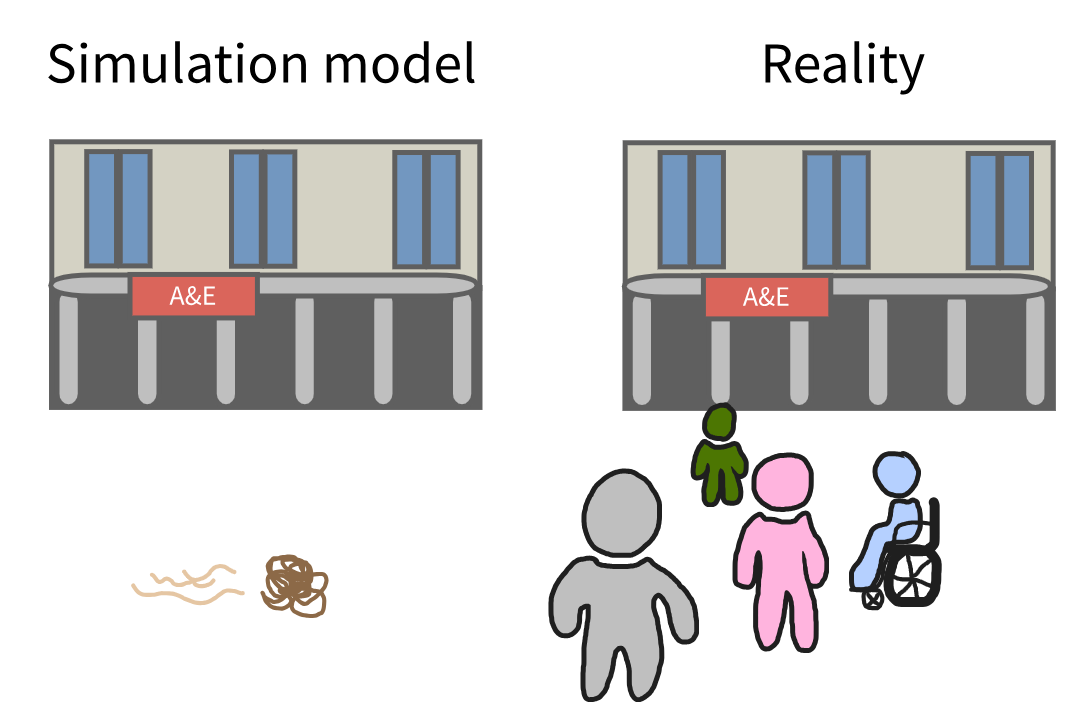
Initialisation bias happens in discrete event simulation when the model starts in an “empty” state that does not match how the real system normally operates.
For systems that naturally begin empty (like a clinic opening each morning), this is fine. However, for many systems that are usually already busy, starting empty skews the results at the beginning of a run. For example, making waiting times, queue lengths or resource use look lower than they would be in reality.
It is important to address this bias as part of your model experimentation validation (see verification and validation page). There are two main strategies for addressing initialisation bias:
- Initial conditions: Start the model with some entities or resources already presents (e.g., a certain number of patients already waiting). This can be difficult though, as it requires relevant data for determining an appropriate starting state.
- Warm-up periods: Run the model for a while until it reaches a steady state, then begin collecting results from that point onwards.
Since finding suitable initial conditions can be complicated, this book will focus on the warm-up method.
Adding a warm-up period to the model
Parameter class
The run_length parameter has been replaced by two new parameters:
warm_up_perioddata_collection_period
class Parameters:
"""
Parameter class.
Attributes
----------
interarrival_time : float
Mean time between arrivals (minutes).
consultation_time : float
Mean length of doctor's consultation (minutes).
number_of_doctors : int
Number of doctors.
warm_up_period : int
Duration of the warm-up period (minutes).
data_collection_period : int
Duration of the data collection period (minutes).
verbose : bool
Whether to print messages as simulation runs.
"""
def __init__(
self, interarrival_time=5, consultation_time=10,
number_of_doctors=3, warm_up_period=30, data_collection_period=40,
verbose=True
):
"""
Initialise Parameters instance.
Parameters
----------
interarrival_time : float
Time between arrivals (minutes).
consultation_time : float
Length of consultation (minutes).
number_of_doctors : int
Number of doctors.
warm_up_period : int
Duration of the warm-up period (minutes).
data_collection_period : int
Duration of the data collection period (minutes).
verbose : bool
Whether to print messages as simulation runs.
"""
self.interarrival_time = interarrival_time
self.consultation_time = consultation_time
self.number_of_doctors = number_of_doctors
self.warm_up_period = warm_up_period
self.data_collection_period = data_collection_period
self.verbose = verbosePatient class
The new attribute period stores a marker which we can use in print() messages to more easily distinguish patients who arrive in the warm-up (🔸 WU) v.s. the data collection period (🔹 DC).
class Patient:
"""
Represents a patient.
Attributes
----------
patient_id : int
Unique patient identifier.
period : str
Arrival period (warm up or data collection) with emoji.
arrival_time : float
Time patient entered the system (minutes).
"""
def __init__(self, patient_id, period, arrival_time):
"""
Initialises a new patient.
Parameters
----------
patient_id : int
Unique patient identifier.
period : str
Arrival period (warm up or data collection) with emoji.
arrival_time : float
Time patient entered the system (minutes).
"""
self.patient_id = patient_id
self.period = period
self.arrival_time = arrival_timeModel class
The changes to Model are explained below…
class Model:
"""
Simulation model.
Attributes
----------
param : Parameters
Simulation parameters.
run_number : int
Run number for random seed generation.
env : simpy.Environment
The SimPy environment for the simulation.
doctor : simpy.Resource
SimPy resource representing doctors.
patients : list
List of Patient objects.
arrival_dist : Exponential
Distribution used to generate random patient inter-arrival times.
consult_dist : Exponential
Distribution used to generate length of a doctor's consultation.
"""
def __init__(self, param, run_number):
"""
Create a new Model instance.
Parameters
----------
param : Parameters
Simulation parameters.
run_number : int
Run number for random seed generation.
"""
self.param = param
self.run_number = run_number
# Create SimPy environment
self.env = simpy.Environment()
# Create resource
self.doctor = simpy.Resource(
self.env, capacity=self.param.number_of_doctors
)
# Create a random seed sequence based on the run number
ss = np.random.SeedSequence(self.run_number)
seeds = ss.spawn(2)
# Set up attributes to store results
self.patients = []
# Initialise distributions
self.arrival_dist = Exponential(mean=self.param.interarrival_time,
random_seed=seeds[0])
self.consult_dist = Exponential(mean=self.param.consultation_time,
random_seed=seeds[1])
def generate_arrivals(self):
"""
Process that generates patient arrivals.
"""
while True:
# Sample and pass time to next arrival
sampled_iat = self.arrival_dist.sample()
yield self.env.timeout(sampled_iat)
# Check whether arrived during warm-up or data collection
if self.env.now < self.param.warm_up_period:
period = "\U0001F538 WU"
else:
period = "\U0001F539 DC"
# Create a new patient
patient = Patient(patient_id=len(self.patients)+1,
period=period,
arrival_time=self.env.now)
self.patients.append(patient)
# Print arrival time
if self.param.verbose:
print(f"{patient.period} Patient {patient.patient_id} " +
f"arrives at time: {patient.arrival_time:.3f}")
# Start process of consultation
self.env.process(self.consultation(patient))
def consultation(self, patient):
"""
Process that simulates a consultation.
Parameters
----------
patient :
Instance of the Patient() class representing a single patient.
"""
# Patient requests access to a doctor (resource)
with self.doctor.request() as req:
yield req
if self.param.verbose:
print(f"{patient.period} Patient {patient.patient_id} " +
f"starts consultation at: {self.env.now:.3f}")
# Sample consultation duration and pass time spent with doctor
time_with_doctor = self.consult_dist.sample()
yield self.env.timeout(time_with_doctor)
# Record end time
if self.param.verbose:
print(f"{patient.period} Patient {patient.patient_id} " +
f"leaves at: {self.env.now:.3f}")
def reset_results(self):
"""
Reset results.
"""
self.patients = []
def warmup(self):
"""
Reset result collection after the warm-up period.
"""
if self.param.warm_up_period > 0:
# Delay process until warm-up period has completed
yield self.env.timeout(self.param.warm_up_period)
# Reset results variables
self.reset_results()
if self.param.verbose:
print(f"Warm up period ended at time: {self.env.now}")
def run(self):
"""
Run the simulation.
"""
# Schedule arrival generator and warm-up
self.env.process(self.generate_arrivals())
self.env.process(self.warmup())
# Run the simulation
self.env.run(until=(self.param.warm_up_period +
self.param.data_collection_period))Explaining changes to the Model class…
# Check whether arrived during warm-up or data collection
if self.env.now < self.param.warm_up_period:
period = "\U0001F538 WU"
else:
period = "\U0001F539 DC"
# Create a new patient
patient = Patient(patient_id=len(self.patients)+1,
period=period,
arrival_time=self.env.now)
self.patients.append(patient)
# Print arrival time
if self.param.verbose:
print(f"{patient.period} Patient {patient.patient_id} arrives " +
f"at time: {patient.arrival_time:.3f}")
...
if self.param.verbose:
print(f"{patient.period} Patient {patient.patient_id} " +
f"starts consultation at: {self.env.now:.3f}")
...
if self.param.verbose:
print(f"{patient.period} Patient {patient.patient_id} " +
f"leaves at: {self.env.now:.3f}")When a patient arrives, we now check whether this occurred during the warm-up period (self.env.now < self.param.warm_up_period). Each patient is then labelled as either warm-up (🔸 WU) or data collection (🔹 DC), stored as an attribute.
The label is used when printing times, helpful for debugging and verifying event sequencing.
def reset_results(self):
"""
Reset results.
"""
self.patients = []The reset_results() method clears the self.patients list once the length of the warm-up period has elapsed, so that arrivals during that time do not contribute to the results.
def warmup(self):
"""
Reset result collection after the warm-up period.
"""
if self.param.warm_up_period > 0:
# Delay process until warm-up period has completed
yield self.env.timeout(self.param.warm_up_period)
# Reset results variables
self.reset_results()
if self.param.verbose:
print(f"Warm up period ended at time: {self.env.now}")The warmup() process waits until the length of the warm-up period has elapsed, and then calls reset_results() to clear patient records. This ensures that data collection starts with a clean slate.
def run(self):
"""
Run the simulation.
"""
# Schedule arrival generator and warm-up
self.env.process(self.generate_arrivals())
self.env.process(self.warmup())
# Run the simulation
self.env.run(until=(self.param.warm_up_period +
self.param.data_collection_period))In the run() method, there were two changes:
- Adding
warmup()as a SimPy process usingself.env.process(). - The total run time is now the sum of
warm_up_periodanddata_collection_period.
Run the model
param = Parameters()
model = Model(param=param, run_number=0)
model.run()🔸 WU Patient 1 arrives at time: 16.468
🔸 WU Patient 1 starts consultation at: 16.468
🔸 WU Patient 2 arrives at time: 20.283
🔸 WU Patient 2 starts consultation at: 20.283
🔸 WU Patient 2 leaves at: 24.103
🔸 WU Patient 1 leaves at: 24.499
🔸 WU Patient 3 arrives at time: 26.545
🔸 WU Patient 3 starts consultation at: 26.545
🔸 WU Patient 4 arrives at time: 27.675
🔸 WU Patient 4 starts consultation at: 27.675
🔸 WU Patient 5 arrives at time: 28.779
🔸 WU Patient 5 starts consultation at: 28.779
Warm up period ended at time: 30
🔸 WU Patient 3 leaves at: 30.187
🔸 WU Patient 4 leaves at: 32.970
🔹 DC Patient 1 arrives at time: 37.778
🔹 DC Patient 1 starts consultation at: 37.778
🔹 DC Patient 2 arrives at time: 38.108
🔹 DC Patient 2 starts consultation at: 38.108
🔹 DC Patient 3 arrives at time: 42.611
🔹 DC Patient 4 arrives at time: 44.088
🔹 DC Patient 2 leaves at: 47.598
🔹 DC Patient 3 starts consultation at: 47.598
🔹 DC Patient 5 arrives at time: 55.588
🔸 WU Patient 5 leaves at: 56.662
🔹 DC Patient 4 starts consultation at: 56.662
🔹 DC Patient 1 leaves at: 57.389
🔹 DC Patient 5 starts consultation at: 57.389
🔹 DC Patient 4 leaves at: 62.537
🔹 DC Patient 6 arrives at time: 64.880
🔹 DC Patient 6 starts consultation at: 64.880
🔹 DC Patient 7 arrives at time: 64.954Parameter function
The run_length parameter has been replaced by two new parameters:
warm_up_perioddata_collection_period
Also, verbose is now set as FALSE as warmup we dont do on those we do on results so focusing on that
#' Generate parameter list.
#'
#' @param interarrival_time Numeric. Time between arrivals (minutes).
#' @param consultation_time Numeric. Mean length of doctor's
#' consultation (minutes).
#' @param number_of_doctors Numeric. Number of doctors.
#' @param warm_up_period Numeric. Duration of the warm-up period (minutes).
#' @param data_collection_period Numeric. Duration of the data collection
#' period (minutes).
#' @param verbose Boolean. Whether to print messages as simulation runs.
#'
#' @return A named list of parameters.
#' @export
create_params <- function(
interarrival_time = 5L,
consultation_time = 10L,
number_of_doctors = 3L,
warm_up_period = 30L,
data_collection_period = 40L,
verbose = FALSE
) {
list(
interarrival_time = interarrival_time,
consultation_time = consultation_time,
number_of_doctors = number_of_doctors,
warm_up_period = warm_up_period,
data_collection_period = data_collection_period,
verbose = verbose
)
}Model function
Add get_mon_arrivals() and get_mon_resources().
#' Run simulation.
#'
#' @param param List. Model parameters.
#' @param run_number Numeric. Run number for random seed generation.
#'
#' @importFrom simmer add_generator add_resource get_mon_arrivals
#' @importFrom simmer get_mon_resources release run seize simmer timeout
#' @importFrom simmer trajectory
#'
#' @return Named list with tables `arrivals` and `resources`.
#' @export
model <- function(param, run_number) {
# Set random seed based on run number
set.seed(run_number)
# Create simmer environment
env <- simmer("simulation", verbose = param[["verbose"]])
# Define the patient trajectory
patient <- trajectory("consultation") |>
seize("doctor", 1L) |>
timeout(function() {
rexp(n = 1L, rate = 1L / param[["consultation_time"]])
}) |>
release("doctor", 1L)
env <- env |>
# Add doctor resource
add_resource("doctor", param[["number_of_doctors"]]) |>
# Add patient generator
add_generator("patient", patient, function() {
rexp(n = 1L, rate = 1L / param[["interarrival_time"]])
}) |>
# Run the simulation
simmer::run(until = (param[["warm_up_period"]] +
param[["data_collection_period"]]))
# Extract information on arrivals and resources from simmer environment
result <- list(
arrivals = get_mon_arrivals(env, per_resource = TRUE, ongoing = TRUE),
resources = get_mon_resources(env)
)
result
}Run the model
We now get two tables returned:
arrivalswhich records the start, end and activity time of each patient with each resource.resourceswhich records when the doctor resources transition between being available and unavailable.
param <- create_params()
result <- model(param = param, run_number = 1L)
result$arrivals
name start_time end_time activity_time resource replication
1 patient0 3.775909 5.232976 1.457067 doctor 1
2 patient1 9.684123 14.044809 4.360686 doctor 1
3 patient2 10.383099 22.678720 12.295621 doctor 1
4 patient3 24.857942 34.423617 9.565675 doctor 1
5 patient5 28.291586 40.667622 12.376036 doctor 1
6 patient4 27.556356 41.463708 13.907351 doctor 1
7 patient6 32.101735 44.969049 10.545432 doctor 1
8 patient8 59.397626 62.766961 3.369335 doctor 1
9 patient10 65.613758 68.554962 2.941204 doctor 1
10 patient11 68.823221 69.883947 1.060726 doctor 1
11 patient9 62.671359 NA NA doctor 1
12 patient7 54.221407 NA NA doctor 1
$resources
resource time server queue capacity queue_size system limit replication
1 doctor 3.775909 1 0 3 Inf 1 Inf 1
2 doctor 5.232976 0 0 3 Inf 0 Inf 1
3 doctor 9.684123 1 0 3 Inf 1 Inf 1
4 doctor 10.383099 2 0 3 Inf 2 Inf 1
5 doctor 14.044809 1 0 3 Inf 1 Inf 1
6 doctor 22.678720 0 0 3 Inf 0 Inf 1
7 doctor 24.857942 1 0 3 Inf 1 Inf 1
8 doctor 27.556356 2 0 3 Inf 2 Inf 1
9 doctor 28.291586 3 0 3 Inf 3 Inf 1
10 doctor 32.101735 3 1 3 Inf 4 Inf 1
11 doctor 34.423617 3 0 3 Inf 3 Inf 1
12 doctor 40.667622 2 0 3 Inf 2 Inf 1
13 doctor 41.463708 1 0 3 Inf 1 Inf 1
14 doctor 44.969049 0 0 3 Inf 0 Inf 1
15 doctor 54.221407 1 0 3 Inf 1 Inf 1
16 doctor 59.397626 2 0 3 Inf 2 Inf 1
17 doctor 62.671359 3 0 3 Inf 3 Inf 1
18 doctor 62.766961 2 0 3 Inf 2 Inf 1
19 doctor 65.613758 3 0 3 Inf 3 Inf 1
20 doctor 68.554962 2 0 3 Inf 2 Inf 1
21 doctor 68.823221 3 0 3 Inf 3 Inf 1
22 doctor 69.883947 2 0 3 Inf 2 Inf 1Warm-up function
To implement a warm-up period in simmer, we need to modify these output dataframes. We have produced a function to do this - filter_warmup():
#' Filters arrivals and resources to remove warm-up results.
#'
#' @param result Named list with two tables: monitored arrivals & resources.
#' @param warm_up_period Length of warm-up period.
#'
#' @importFrom dplyr arrange filter group_by mutate n slice ungroup
#' @importFrom rlang .data
#'
#' @return The named list `result`, but with the tables (`arrivals` and
#' `resources`) filtered to remove warm-up results.
#' @export
filter_warmup <- function(result, warm_up_period) {
# Skip filtering if the warm-up period is zero
if (warm_up_period == 0L) return(result)
# Arrivals: Keep only patients who came during the data collection period
result[["arrivals"]] <- result[["arrivals"]] |>
group_by(.data[["name"]]) |>
filter(all(.data[["start_time"]] >= warm_up_period)) |>
ungroup()
# Resources: Filter to resource events in the data collection period
dc_resources <- filter(result[["resources"]],
.data[["time"]] >= warm_up_period)
if (nrow(dc_resources) > 0L) {
# For each resource, get the last even during warm-up, and replace time
# for that event with the start time of the data collection period
last_usage <- result[["resources"]] |>
filter(.data[["time"]] < warm_up_period) |>
arrange(.data[["time"]]) |>
group_by(.data[["resource"]]) |>
slice(n()) |>
mutate(time = warm_up_period) |>
ungroup()
# Set that last event of the resource/s as the first row of the dataframe
# else calculations would assume all resources were idle at the start of
# the data collection period
result[["resources"]] <- rbind(last_usage, dc_resources)
} else {
# No events after warm-up; use filtered resource table as is
result[["resources"]] <- dc_resources
}
result
}Explaining the warm-up function…
#' Filters arrivals and resources to remove warm-up patients.
#'
#' @param result Named list with two tables: monitored arrivals & resources.
#' @param warm_up_period Length of warm-up period.
#'
#' @importFrom dplyr arrange filter group_by mutate n slice ungroup
#' @importFrom rlang .data
#'
#' @return The named list `result`, but with the tables (`arrivals` and
#' `resources`) filtered to remove warm-up patients.
#' @export
filter_warmup <- function(result, warm_up_period) {The function accepts two inputs: result and warm_up_period.
# Skip filtering if the warm-up period is zero
if (warm_up_period == 0L) return(result)If warm_up_period is zero, the function instantly returns the original results list unchanged, as no filtering is needed.
# Arrivals: Keep only patients who came during the data collection period
result[["arrivals"]] <- result[["arrivals"]] |>
group_by(.data[["name"]]) |>
filter(all(.data[["start_time"]] >= warm_up_period)) |>
ungroup()The arrivals table is filtered to remove patients who arrived during the warm-up period (even if they have activity times after warm-up).
# Resources: Filter to resource events in the data collection period
dc_resources <- filter(result[["resources"]],
.data[["time"]] >= warm_up_period)The resources table is filtered so it only includes events that occurred after the warm-up period.
if (nrow(dc_resources) > 0L) {
# For each resource, get the last even during warm-up, and replace time
# for that event with the start time of the data collection period
last_usage <- result[["resources"]] |>
filter(.data[["time"]] < warm_up_period) |>
arrange(.data[["time"]]) |>
group_by(.data[["resource"]]) |>
slice(n()) |>
mutate(time = warm_up_period) |>
ungroup()
# Set that last event of the resource/s as the first row of the dataframe
# else calculations would assume all resources were idle at the start of
# the data collection period
result[["resources"]] <- rbind(last_usage, dc_resources)
} else {
# No events after warm-up; use filtered resource table as is
result[["resources"]] <- dc_resources
}
return(result)The resources table requires an extra change: we add a row for each resource representing their last known state (i.e., number of resources occupied) from the warm-up period, settings its timestamp to the start of data collection.
Without this step, any statistics (e.g., utilisation) would falsely assume the resources were unused at the beginning of the data collection window. This adjustments preserves the true initial state of each resource at that timepoint.
Run filter_warmup()
Our warm-up period is 30:
param[["warm_up_period"]][1] 30We can see that both tables now have no start_time or time before 30.
We can also see the first row in resources setting the state at the start of the data collection period.
filter_warmup(result = result, warm_up_period = param[["warm_up_period"]])$arrivals
# A tibble: 6 × 6
name start_time end_time activity_time resource replication
<chr> <dbl> <dbl> <dbl> <chr> <int>
1 patient6 32.1 45.0 10.5 doctor 1
2 patient8 59.4 62.8 3.37 doctor 1
3 patient10 65.6 68.6 2.94 doctor 1
4 patient11 68.8 69.9 1.06 doctor 1
5 patient9 62.7 NA NA doctor 1
6 patient7 54.2 NA NA doctor 1
$resources
# A tibble: 14 × 9
resource time server queue capacity queue_size system limit replication
<chr> <dbl> <int> <int> <dbl> <dbl> <int> <dbl> <int>
1 doctor 30 3 0 3 Inf 3 Inf 1
2 doctor 32.1 3 1 3 Inf 4 Inf 1
3 doctor 34.4 3 0 3 Inf 3 Inf 1
4 doctor 40.7 2 0 3 Inf 2 Inf 1
5 doctor 41.5 1 0 3 Inf 1 Inf 1
6 doctor 45.0 0 0 3 Inf 0 Inf 1
7 doctor 54.2 1 0 3 Inf 1 Inf 1
8 doctor 59.4 2 0 3 Inf 2 Inf 1
9 doctor 62.7 3 0 3 Inf 3 Inf 1
10 doctor 62.8 2 0 3 Inf 2 Inf 1
11 doctor 65.6 3 0 3 Inf 3 Inf 1
12 doctor 68.6 2 0 3 Inf 2 Inf 1
13 doctor 68.8 3 0 3 Inf 3 Inf 1
14 doctor 69.9 2 0 3 Inf 2 Inf 1Explore the example models
Nurse visit simulation
Click to visit pydesrap_mms repository
| Key file | simulation/param.pysimulation/model.py |
| What to look for? | The model has a 27-day warm-up period. The implementation of a warm-up period is like our example above, with a warm_up_complete() function that triggers a function init_results_variables() to clear results when the warm-up period has passed. |
Click to visit rdesrap_mms repository
| Key file | R/parameters.RR/model.RR/warmup.R |
| What to look for? | The model has a 27-day warm-up period. The implementation is consistent with above - with a filter_warmup() function applied to the results. |
Stroke pathway simulation
Click to visit pydesrap_stroke repository
| Key file | simulation/parameters.pysimulation/model.py |
| What to look for? | The model has a 3-year warm-up period. The implementation is again consistent with above - with a warm_up() function that triggers the function reset_results() to clear results when the warm-up period has passed. |
Click to visit rdesrap_stroke repository
| Key file | R/parameters.RR/model.RR/warmup.R |
| What to look for? | The model has a 3-year warm-up period. The implementation of the warm-up period is simpler than above, as this model only uses the dataframe from get_mon_arrivals() (and not get_mon_resources()), so the filter_warmup() function is much shorter! |
Test yourself
If you haven’t already, try adding a warm-up period to your model.
Task:
Copy over the modifications above and run the model.
Try changing the
warm_up_periodanddata_collection_periodparameters - how does this affect which patients are included in results?