#### 1. Installation ###########
#### This code has been created using R version 4.3.1
#### All packages used by this model are provided here
#### Comment out the below section which installs the relevant packages after the first run of the model
# install.packages("shiny", quiet = TRUE) ### the quiet argument is used to avoid warnings appearing in the console (useful for later conversion to web app)
# install.packages("gtools", quiet = TRUE)
# install.packages("openxlsx", quiet = TRUE)
# install.packages("flexsurv", quiet = TRUE)
# install.packages("tidyverse", quiet = TRUE)
# install.packages("data.table", quiet = TRUE)
# install.packages("heemod", quiet = TRUE)
# install.packages("logOfGamma", quiet = TRUE)
# install.packages("ggplot2", quiet = TRUE)
# install.packages("survminer", quiet = TRUE)
# install.packages("officer", quiet = TRUE)
# install.packages("officedown", quiet = TRUE)
# install.packages("magrittr", quiet = TRUE)
# install.packages("Hmisc", quiet = TRUE)
# install.packages("future.apply", quiet = TRUE)
# install.packages("crosstable", quiet = TRUE)
# install.packages("flextable", quiet = TRUE)
# install.packages("stringr", quiet = TRUE)
# install.packages("BCEA", quiet = TRUE)
# install.packages("collapse", quiet = TRUE)
# install.packages("scales", quiet = TRUE)
# install.packages("Matrix", quiet = TRUE)
# install.packages("dplyr", quiet = TRUE)
# install.packages("progressr", quiet = TRUE)
# install.packages("microbenchmark", quiet = TRUE)
### Loading libraries
#### This section needs to be run every time and calls each package from the library
library(shiny, quiet = TRUE)
library(gtools, quiet = TRUE)
library(openxlsx, quiet = TRUE)
library(flexsurv, quiet = TRUE)
library(tidyverse, quiet = TRUE)
library(data.table, quiet = TRUE)
library(heemod, quiet = TRUE)
library(logOfGamma, quiet = TRUE)
library(ggplot2, quiet = TRUE)
library(survminer, quiet = TRUE)
library(officer, quiet = TRUE)
library(officedown, quiet = TRUE)
library(magrittr, quiet = TRUE)
library(Hmisc, quiet = TRUE)
library(future.apply, quiet = TRUE)
library(crosstable, quiet = TRUE)
library(flextable, quiet = TRUE)
library(stringr, quiet = TRUE)
library(BCEA, quiet = TRUE)
library(collapse, quiet = TRUE)
library(scales, quiet = TRUE)
library(Matrix, quiet = TRUE)
library(dplyr, quiet = TRUE)
library(progressr, quiet = TRUE)
library(microbenchmark, quiet = TRUE)
library(knitr, quiet = TRUE)Set-up
This page contains some of the basic set-up steps like loading the functions and lots of the model inputs, as well as establishing the possible treatment sequences.
Load required packages
Set file paths
# Set path to folders
d_path = "../../../1_Data"
s_path = "../../../2_Scripts"
f_path = "../../../3_Functions"
o_path = "../../../4_Output"Define some of the model settings
The majority of this code block is dedicated to setting the model to run sequentially or in parallel. If running the base case using Model_Structure.R as provided (e.g. with pre-run survival analysis), you may find that the quickest option is to run the model sequentially. This is already set up by default, with keep_free_cores <- NA. For reference, the run time for this on an Intel Core i7-12700H with 32GB RAM running Ubuntu 22.04.4 Linux was 40 minutes.
# Multi-core processing:
#
# Instructions.
#
# This model is highly RAM intensive. You need a lot of RAM on your computer
# to run this model due to the large amount of very large matrix multiplications
# (up to approximately 15,000 discrete health states in the model). Therefore,
# in order to efficiently run the model, it is a balancing act between RAM
# usage and CPU usage.
#
# Some rough guidance is:
#
# - If you have 8GB of RAM on your computer, you can run this model with 2 cores only
# but it may even be faster to run in series if you have other things open on your
# computer at the same time. Therefore, please set keep_free_cores to NA and run
# the model in series. This is because when the RAM on your computer runs out
# your computer will use the hard-disk instead which is extremely slow.
# - If you have 16GB of RAM on your computer, parallel should be a lot faster.
# On my laptop (I7 8th gen, 16GB RAM, running Linux for low RAM usage) I can
# run with 5 cores whilst using about 12GB of RAM running this model.
# - if you have 24GB or 32GB of RAM, you should be able to run the model with 8
# and up to around 14 cores before running out of RAM whilst running the model.
# - if you are using a HPC, you should be able to run this model with many cores
# due to the typically large amount of RAM available per core in a HPC
#
#
# IF YOU DO NOT WANT MULTICORE SET keep_free_cores TO NA
#
#
keep_free_cores <- NA
if (any(is.na(keep_free_cores), keep_free_cores<0)) {
plan(sequential)
} else {
plan(multisession(workers = max(availableCores()-keep_free_cores,1)))
}The three settings in this code block:
progressr::handlers("progress")- one of the settings for how progress is reported whilst code is runningqc_mode- the input forverboseinf_NMA_AddAssumptionsToNetwork()which, if true, will mean that extra outputs are printed to the console
# Other generic settings for the progress bar and units for table widths
handlers("progress")
#### 2. Loading functions ###########
# This variable is used throughout the model to define whether to provide additional outputs useful for QC or not
# The model will take longer to run when this is set to TRUE
qc_mode <- FALSEImport model functions
The functions for the analysis in Model_Structure.R are stored in the 3_Functions/ folder. Here, they are imported into our environment.
# 2.1. Excel data extraction functions -----------------------------------------
#### These functions are used to extract parameters from the Excel input workbook for use in R
#### During Phase 2 a Shiny front-end will be added to the model which will allow an alternative mechanism to upload these types of inputs
source(file.path(f_path, "excel/extract.R"))
# 2.2. Treatment sequencing functions ----------------------------------------
#### Function: filter to active treatments and lines
##### Takes as an input the defined sequences, evaluation type and line to start the evaluation from
##### Other input is % receiving each subs therapy at each line dependent on previous treatments received
##### Reweights so that the % receiving each treatment sums to 100% within each arm / line being studied
##### Outputs a matrix that has the % receiving each possible combination
source(file.path(f_path, "sequencing/sequences.R"))
# 2.3. Survival analysis functions ---------------------------------------------
# Function: conduct survival analysis
##### by treatment, line, population and outcome fitted survival curves using Flexsurvreg (exp, Weibull, lognormal, loglog, Gompertz, gen gamma)
##### calculation of and adjustment for general population
##### adjustment for treatment effect waning
source(file.path(f_path, "survival/Survival_functions.R"))
source(file.path(f_path, "survival/other_cause_mortality.R"))
source(file.path(f_path, "survival/treatment_effect_waning.R"))
# 2.4 Misc functions ----------------------------------------------------------
### these functions enable smoother data cleaning and manipulation
source(file.path(f_path, "misc/other.R"))
source(file.path(f_path, "misc/shift_and_pad.R"))
source(file.path(f_path, "misc/cleaning.R"))
# 2.4.1 Functions imposing list structures -----------------------------------
source(file.path(f_path, "misc/nesting.R"))
source(file.path(f_path, "misc/discounting.R"))
source(file.path(f_path, "misc/qdirichlet.R"))
source(file.path(f_path, "misc/plotting.R"))
source(file.path(f_path, "misc/structure.R"))
# 2.4.2 Functions calculating HRs from FPNMA coefficients and other FPNMA manipulation ------
source(file.path(f_path, "misc/fpnma_fns.R"))
# 2.5 Utility functions -------------------------------------------------------
source(file.path(f_path, "utility/age_related.R"))
source(file.path(f_path, "costs_and_QALYs/utility_processing.R"))
# 2.6 AE functions --------------------------------------------------------
source(file.path(f_path, "adverse_events/AE_steps.R"))
# 2.7 Cost calculation functions --------------------------------------------
source(file.path(f_path, "costs_and_QALYs/cost_processing.R"))
# 2.8 State transition modelling functions --------------------------------
source(file.path(f_path, "markov/markov.R"))
# 2.9 Patient flow functions ----------------------------------------------
source(file.path(f_path, "patient_flow/overarching.R"))
source(file.path(f_path, "patient_flow/partitioned_survival.R"))
source(file.path(f_path, "patient_flow/markov.R"))
source(file.path(f_path, "patient_flow/drug_costs.R"))
source(file.path(f_path, "patient_flow/hcru_costs.R"))
source(file.path(f_path, "patient_flow/qalys.R"))
source(file.path(f_path, "patient_flow/ae.R"))
# 2.10 Results processing functions ---------------------------------------
source(file.path(f_path, "results/incremental_analysis.R"))
source(file.path(f_path, "results/model_averaging.R"))
source(file.path(f_path, "results/partitioned_survival.R"))
source(file.path(f_path, "misc/severity_modifier.R"))
source(file.path(f_path, "results/results_tables.R"))
source(file.path(f_path, "psa/psa functions.R"))
# 2.11 Office software outputs --------------------------------------------
source(file.path(f_path, "reporting/word_document_output.R"))Get some of the model inputs
Introductory comments
This section of the code is mostly comments that describe:
- The structure of
iwhich contains model inputs - That survival analysis will be conducted using state transition (markov) models and partitioned survival analysis (partSA)
- The five input files, which are detailed within the documentation on the page Input data
# 3. Model inputs structure --------------------------------------------------
# Model inputs should be in a list called i. This list then contains all of the
# inputs for the model, NOT the parameters used to calculate the model. In effect,
# this is a place to store all model information BEFORE it gets boiled down to
# what's needed to run 1 model.
#
# using i allows subsetting by categorisation, which makes things a lot easier
# to find and avoids all long variable names
#
# the structure of i should be by category. There are the following
# categories:
#
# dd - dropdown inputs taken from Excel
# i - parameter inputs taken from Excel
# r_ tables taken from Excel
# List, id and lookup - lists defined and used within the code
# basic - basic inputs (time horizon, cycle length, discount rates, so on so forth)
# surv - survival analysis inputs including raw data
# sequences and seq - inputs and outputs related to the possible sequences of treatments
# cost - drug and hcru costs. All costs are here to keep things together (dosing is not cost)
# util and QALYs - utility and QALY inputs
# misc - misc inputs e.g. graph labelling
#
#### 3.1 Loading input parameters ###########
# This model allows two possible structures to be analysed: state transition with a user definable number of lines
# with health states based on time to discontinuation (drug costs) and progression status (quality of life and movement
# between lines) and PartSA with 3 health states (pre-progression, post-progression and death)
# During Phase 1 of this pilot we use the model to evaluate the decision problem for a single therapy
# (cabo+nivo, defined as molecule 1) starting at 1st line
# During Phase 2 we will adapt this code to evaluate the cost-effectiveness of sequences starting at a user-defined line
# Users can either use the files provided in the repository in which confidential data is redacted by replacing the original numbers with dummy values or, upload their own files using the same format
# The way raw data is fed into the model currently works as follows
# Define the path to where the data file lives using the select file functionality
# The model then processes the file the user selected
# There are a number of files which contain raw or intermediate inputs:
# 1. The Excel user interface - this contains information from company data and the UK RWE
# 2. The proportional hazards NMA CODA RDS file - this contains information from company data
# 3. The fractional polynomials NMA RDS file - this contains information from company data
# 4. Either the raw data file containing the pseudo-IPD for all trials for survival analysis (RWE and company data included); or
# 5. The RDS output from the survival analysis using both RWE and company data
# You will need to manually select the inputs file relevant to your user type, this is not stored on Github as access to CIC information differs by user typeGet inputs from excel workbook and save as list i
Import the file at excel_path using f_excel_extract(), and then tidy i$R_table_param using f_excel_cleanParams().
If the file doesn’t exist, assuming the user is in RStudio, a dialog box will appear with the system files, and the user should then select a file from their directory. The dialog will open in 1_Data/, show only .xlsm files, and the accept/ok button has the text ID6184_RCC_model inputs....xlsm.
Note: The typical naming convention used for this was that variables starting with R_, List_, i_, ui_ or dd_ are likely used within the R scripts. Some of the variables imported from excel are intermediate for calculations within excels, and so - though imported - not necessarily used in the scripts.
# The first part of this code pulls all of the named ranges from the excel workbook, expand the parameters table
#Option to define Excel path on local machine - comment in this and comment out the code below to select file
excel_path <- file.path(d_path, "ID6184_RCC_model inputs FAD version [UK RWE unredacted, ACIC redacted, cPAS redacted].xlsm")
#i <- f_excel_extract(excel_path, verbose = TRUE)
if (file.exists(excel_path)) {
i <- f_excel_extract(excel_path, verbose = FALSE)
} else {
i <- f_excel_extract(rstudioapi::selectFile(
caption = "Select the Excel inputs file (ID6184_RCC_model inputs....xlsm)",
label = "ID6184_RCC_model inputs....xlsm",
path = "./1_Data/",
filter = "Excel Files (*.xlsm)",
existing = TRUE
), verbose = FALSE)
}
i <- c(i,f_excel_cleanParams(i$R_table_param))f_excel_extract() do?
This function uses openxlsx::getNamedRegions() to find named regions in the workbook. These are all stored in a sheet called named ranges. As illustrated in Input data, these consist of two columns:
- A name for the region (e.g.
List_pop1_allowed) - The region, which consists of the: sheet, column/s and row/s (e.g.
=Lists!$BA$11:$BA$22)
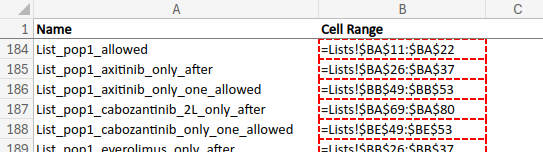
The function f_excel_extract() extracts each of these, saving them into a nested list called i. Each element in the list is a row from the named ranges sheet - for example:
i$List_pop1_allowed [1] "avelumab_plus_axitinib" "axitinib"
[3] "cabozantinib" "everolimus"
[5] "lenvatinib_plus_everolimus" "cabozantinib_plus_nivolumab"
[7] "nivolumab_monotherapy" "pazopanib"
[9] "sunitinib" "tivozanib" The exception is the first element which is a copy of the named ranges sheet:
print(head(i[[1]])) Name Cell.Range
1 apply_waning_to =Lists!$Y$10:$Y$11
2 bc_settings_rng =Lists!$B$99:$B$174
3 cabo_nivo_outcome_from =Lists!$W$10
4 cabo_nivo_outcomes =Lists!$X$10:$X$12
5 count_bc_settings =Lists!$B$97
6 dd_2ndline_NMA ='Model settings'!$G$40f_excel_cleanParams() do?
This function is applied to i$R_table_param which is the full table from the sheet All parameters.
kable(head(i$R_table_param))| Parameter.description | Parameter.name | Mean.current.value |
|---|---|---|
| Include cabo nivo? (1=yes, 0=no) | cabo_nivo_include | 1 |
| Include pem len? (1=yes, 0=no) | pem_len_include | 1 |
| Include panzopanib? (1=yes, 0=no) | pazopanib_include | 1 |
| Include tivozanib? (1=yes, 0=no) | tivozanib_include | 1 |
| Include sunitinib? (1=yes, 0=no) | sunitinib_include | 1 |
| Include cabo monotherapy? (1=yes, 0=no) | cabo_include | 1 |
The function f_excel_cleanParams():
- Converts each row of the table into a list, with value from
Parameter.nameused as the list name - Adds
Meanwhich is simplyMean.current.valueconverted to numeric
These lists were then concatenated with i, so we can access them from i as follows:
i$cabo_nivo_include$Parameter.description
[1] "Include cabo nivo? (1=yes, 0=no)"
$Parameter.name
[1] "cabo_nivo_include"
$Mean.current.value
[1] "1"
$Mean
[1] 1Manually add some extra inputs to i
# Set which decision problem to look at, initially functionality has been geared towards the decision problem for cabozantinib plus nivolumab
i$decision_problem <- "cabo+nivo"
# We then create a place for identifiers. Adding in an object to i full of lookup tables makes automated translation
# possible even when one doesn't know the number of items ex ante, or how they combine.
#
# If the lookup table is correct one can translate id numbers to text strings which are
# consistent throughout the entire model. This is extremely useful as the model can
# be expanded to any number of treatments and potentially even any number of lines
# (up to a reasonable maximum)
i$id <- list(ipd = list())
i$lookup <- list(ipd = list())
# Add distribution names to i
# This model only includes standard parametric distributions as more complex distributions were not deemed to be required for the included treatments
i$distnames <-
c(
gengamma = "gengamma",
exp = "exp",
weibull = "weibull",
lnorm = "lnorm",
gamma = "gamma",
gompertz = "gompertz",
llogis = "llogis"
)Use i to make another list p
The list p is based on i, either copying over parameters or using them to calculate new parameters. The list is first created by f_misc_param_generate_p(). A few extra additions are then made to p in this code block, such as to set a maximum of 4 treatment lines before best supportive care.
# The next step is to then "tidy up" i into another object, p. p doesn't necessarily
# have to house everything, only things that will change in PSA
p <- f_misc_param_generate_p(i)
# Set seed for PSA - note this is done in the script to run the PSA, not here!
# set.seed(1475)
# Max lines within the R model
p$basic$R_maxlines <- 4
# Pass this into p so that p can be used to exclusively compute the model:
p$basic$decision_problem <- i$decision_problemf_misc_param_generate_p() do?
This function consists of relatively simple calculations using parameters from i to generate p. For the full overview of these calculations, check out the code for the function. To give an example though, the function code includes:
p <- list(
basic = list(
th = ceiling(i$ui_time_horizon * 365.25 / 7),
th_y = i$ui_time_horizon,
...
)
...
)In this example, we can see that is makes a copy of i$ui_time_horizon which has time horizon of model in years (40 years) (p$basic$th_y). It also converts the time horizon into weeks (p$basic$th).
i$ui_time_horizon[1] 40p$basic$th[1] 2088p$basic$th_y[1] 40Treatment sequences
Find all possible sequences
The first step in determining the possible treatment sequences is to determine all possible combinations and orders of treatment, saving this as i$sequences.
#### 3.2 Define sequences ###########
#### This code produces a list of possible sequences per population based upon the rules defined for RCC
#### and the user input number of lines
# Add drug names to comparators vector extracted from inputs list.
i$sequences <- f_generate_sequences(
comparators = i$List_comparators,
maxlines = p$basic$R_maxlines
)f_generate_sequences() do?
The input to this function is a list of all the possible treatments (14 options), and the maximum number of treatment lines (4).
i$List_comparators [1] "nivolumab_monotherapy" "cabozantinib_plus_nivolumab"
[3] "nivolumab_plus_ipilimumab" "lenvatinib_plus_pembrolizumab"
[5] "avelumab_plus_axitinib" "pazopanib"
[7] "tivozanib" "sunitinib"
[9] "cabozantinib" "lenvatinib_plus_everolimus"
[11] "everolimus" "axitinib"
[13] "sorafenib" "placebo_BSC" The function f_generate_sequences() outputs a table saved as i$sequences. This contains every possible order and combination of treatments.
kable(head(i$sequences, 3))| V1 | V2 | V3 | V4 | V5 |
|---|---|---|---|---|
| avelumab_plus_axitinib | axitinib | cabozantinib | cabozantinib_plus_nivolumab | BSC |
| avelumab_plus_axitinib | axitinib | cabozantinib | everolimus | BSC |
| avelumab_plus_axitinib | axitinib | cabozantinib | lenvatinib_plus_everolimus | BSC |
dim(i$sequences)[1] 26404 5These varied from a single treatment to up to four subsequent treatments, but always ended with best supportive case (BSC).
kable(head(i$sequences[i$sequences$V3=="",], 3))| V1 | V2 | V3 | V4 | V5 |
|---|---|---|---|---|
| avelumab_plus_axitinib | BSC | |||
| axitinib | BSC | |||
| cabozantinib | BSC |
Filter to valid sequences
The table of all possible treatment sequences is then filtered down to valid sequences (for example, removing drugs if not allowed for a given population, or after another particular drug).
# restrict the pathways to those that are possible and permitted.
i$sequences <- as.data.frame(i$sequences)
populations <- i$i_nr_populations
seqs <- NULL
invisible(
for (population in 1:populations) {
cat("Applying sequence restrictions to population", population,"\n")
s <- f_path_tx_restrict(
sequences = i$sequences,
allowed = f_get_allowed_lists(i, population), #overall list of allowed drugs in this popn
L1 = f_get_L1_lists(i, population), # 1L drugs allowed in this popn
L2 = f_get_L2_lists(i, population), # 2L drugs allowed in this popn
L3 = f_get_L3_lists(i, population), # 3L drugs allowed in this popn
L4 = f_get_L4_lists(i, population), # 4L drugs allowed in this popn
only_after = f_get_only_after_lists(i, population), #list of restrictions where tx can be only after the listed txs
not_immediate_after = f_get_not_immediate_after_lists(i, population), #list of restrictions where tx can be only immediately before the listed txs
one_in_list = f_get_one_in_list_lists(i, population), #list of restrictions where only one of the tx in each list is allowed
only_after_one = f_get_only_after_one_lists(i, population), #list of restrictions where only one of the listed treatments is allowed prior to current therapy
L2_only_after = f_get_2L_only_after_lists(i, population), #list of 2L+ restrictions: if drug is used 2L, 3L or 4L, can only be after drug x
L2_only_immediate_after = f_get_2L_only_immediate_after_lists(i, population), #list of 2L+ restrictions: if drug is used 2L, 3L or 4L, can only be immediately after drug x
L2_only_one = f_get_2L_only_one_lists(i, population) #list of 2L+ drugs where only one of them allowed in a given sequence
)
s <- cbind(rep(paste0("pop", population),nrow(s)), s)
colnames(s) <- paste0('V', seq_len(ncol(s))) # rbind no longer likes un-named columns so added this
seqs <- rbind(seqs, s)
}
)
rownames(seqs) <- NULL
i$sequences <- seqs
#### Uncomment this code to view the sequences and write the sequences defined to csv
# i$sequences
# write.csv(seqs, "4_Output/sequences.csv", row.names = F)
rm(s, seqs, populations)
# define number of cycles and a vector of the cycles This code chunk restricted to valid sequences by population. In this analysis, there are four populations defined by time since an immuno-oncology (IO) treatment, and International Metastatic Renal Cell Carcinoma Database Consortium (IMDC) risk status. They are:
- pop1 >12m since IO, favourable risk
- pop2 >12m since IO, intermediate/poor risk
- pop3 <12m since IO, favourable risk
- pop4 <12m since IO, intermediate/poor risk
Each risk group has to be broken down by time since IO as there are five treatments that cannot be used within 12 months of an adjuvant IO treatment (4.3.5.6 in Assessment Report [1]). An adjuvant treatment is one given alongside the primary treatment.
By population and line
For each population, there are a list of valid treatments at each line of therapy (first-line through to fourth). For example, valid first-line treatments for population 1 are:
# Pop1 1L treatments
f_get_L1_lists(i, 1)[1] "avelumab_plus_axitinib" "cabozantinib_plus_nivolumab"
[3] "pazopanib" "sunitinib"
[5] "tivozanib" Only after
There are some treatments that can only come after other treatments. For example, for population 1:
- Axitinib can only be administered after a tyrosine kinase inhibitor (TKI) or cytokine treatment
- Everolimus can only be administered after a vascular endothelial growth factor (VEGF) treatment
- At 2L 3L or 4L, cabozantinib can only be administered after one of the listed treatments
f_get_only_after_lists(i, 1)$axitinib
[1] "avelumab_plus_axitinib" "cabozantinib"
[3] "lenvatinib_plus_everolimus" "cabozantinib_plus_nivolumab"
[5] "pazopanib" "lenvatinib_plus_pembrolizumab"
[7] "sunitinib" "tivozanib"
$everolimus
[1] "avelumab_plus_axitinib" "axitinib"
[3] "cabozantinib" "lenvatinib_plus_everolimus"
[5] "cabozantinib_plus_nivolumab" "pazopanib"
[7] "lenvatinib_plus_pembrolizumab" "sunitinib"
[9] "tivozanib" f_get_2L_only_after_lists(i, 1)$cabozantinib
[1] "avelumab_plus_axitinib" "axitinib"
[3] "lenvatinib_plus_everolimus" "cabozantinib_plus_nivolumab"
[5] "pazopanib" "lenvatinib_plus_pembrolizumab"
[7] "sunitinib" "tivozanib" Not immediately after
Some treatments cannot come immediately after another treatment. For example, for population 1:
- Lenvatinib plus everolimus must not come immediately after nivolumab plus ipilimumab
- At 2L 3L or 4L, pazopanib and sunitinib and tivozanib must not come immediately after their respective listed treatments
f_get_not_immediate_after_lists(i, 1)$lenvatinib_plus_everolimus
[1] "nivolumab_plus_ipilimumab"f_get_2L_only_immediate_after_lists(i, 1)$pazopanib
[1] "avelumab_plus_axitinib" "lenvatinib_plus_pembrolizumab"
[3] "cabozantinib_plus_nivolumab" "nivolumab_plus_ipilimumab"
$sunitinib
[1] "avelumab_plus_axitinib" "lenvatinib_plus_pembrolizumab"
[3] "cabozantinib_plus_nivolumab" "nivolumab_plus_ipilimumab"
$tivozanib
[1] "avelumab_plus_axitinib" "lenvatinib_plus_pembrolizumab"
[3] "cabozantinib_plus_nivolumab" "nivolumab_plus_ipilimumab" Only one from list
Some treatments are not allowed if another has already been given at any point prior.
For example, for population 1 there are five lists of treatments where only one treatment from each list is allowed.
There are no restrictions like this for population 1 specific to just 2L 3L or 4L treatments (hence, empty list).
f_get_one_in_list_lists(i, 1)$axitinib
[1] "avelumab_plus_axitinib" "axitinib"
$cabozantinib
[1] "cabozantinib" "cabozantinib_plus_nivolumab"
$everolimus
[1] "lenvatinib_plus_everolimus" "everolimus"
$io
[1] "avelumab_plus_axitinib" "nivolumab_plus_ipilimumab"
[3] "cabozantinib_plus_nivolumab" "nivolumab_monotherapy"
[5] "lenvatinib_plus_pembrolizumab"
$nivolumab
[1] "nivolumab_plus_ipilimumab" "cabozantinib_plus_nivolumab"
[3] "nivolumab_monotherapy"
$TKIs
[1] "sunitinib" "pazopanib" "tivozanib"f_get_2L_only_one_lists(i, 1)named list()Only one allowed before
In other cases, a treatment is not allowed if more than one of a particular category of treatment has been given. For example, for population 1:
- Of the listed treatments, only one is allowed before lenvatinib plus everolimus
f_get_only_after_one_lists(i, 1)$lenvatinib_plus_everolimus
[1] "avelumab_plus_axitinib" "axitinib"
[3] "cabozantinib" "cabozantinib_plus_nivolumab"
[5] "pazopanib" "lenvatinib_plus_pembrolizumab"
[7] "sunitinib" "tivozanib" f_path_tx_restrict() do?
The function f_path_tx_restrict() is defined in sequences.R (there is also a function of the same name in rccFunctions.R but this is not sourced).
Its purpose is to restrict the table of all possible sequences to just the valid sequences for each population, restricting it from 26404 rows with possible treatment sequences to just 744.
dim(i$sequences)[1] 26404 5The inputs to this function are lists defining valid treatments by different criteria (e.g. line of therapy, subsequent treatments), as detailed in the note above.
Within f_path_tx_restrict(), there are then several other functions which take these lists and use them to remove invalid sequences from the table.
For example, the allowed treatments identified using f_get_allowed_lists() are input to f_path_tx_restrict() as allowed. The function f_path_allowed() then uses that list to remove invalid drugs for a given population:
s <- f_path_allowed(s, allowed[[1]])Looking at an excerpt of the code for f_path_allowed(), we can see that is adds “BSC” and no treatment (““) as valid options, and then only keeps rows if their treatments are in (%in%) the list of valid treatments.
rule <- c(rule, "BSC", "")
for (n in 1:ncol(perms)) {
perms <- perms[perms[,n] %in% rule,]
}Applying sequence restrictions to population 1
Dropping drugs not allowed for this population.
applying rule: avelumab_plus_axitinib axitinib cabozantinib everolimus lenvatinib_plus_everolimus cabozantinib_plus_nivolumab nivolumab_monotherapy pazopanib sunitinib tivozanib are only allowed treatments.
Permutations before applying rule: 26404
Permutations after applying rule : 5860
applying rule: drug line restrictions.
Permutations before applying rule: 5860
Permutations after applying rule : 628
[1] "axitinib"
applying rule. axitinib is only allowed after avelumab_plus_axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 628
Permutations after applying rule : 628
[1] "everolimus"
applying rule. everolimus is only allowed after avelumab_plus_axitinib axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 628
Permutations after applying rule : 628
[1] "lenvatinib_plus_everolimus"
applying rule. lenvatinib_plus_everolimus is not allowed immediately after nivolumab_plus_ipilimumab
Permutations before applying rule: 628
Permutations after applying rule : 628
[1] "axitinib"
applying rule axitinib : avelumab_plus_axitinib axitinib cannot be in one permutation
Permutations before applying rule: 628
Permutations after applying rule : 559
[1] "cabozantinib"
applying rule cabozantinib : cabozantinib cabozantinib_plus_nivolumab cannot be in one permutation
Permutations before applying rule: 559
Permutations after applying rule : 520
[1] "everolimus"
applying rule everolimus : lenvatinib_plus_everolimus everolimus cannot be in one permutation
Permutations before applying rule: 520
Permutations after applying rule : 452
[1] "io"
applying rule io : avelumab_plus_axitinib nivolumab_plus_ipilimumab cabozantinib_plus_nivolumab nivolumab_monotherapy lenvatinib_plus_pembrolizumab cannot be in one permutation
Permutations before applying rule: 452
Permutations after applying rule : 400
[1] "nivolumab"
applying rule nivolumab : nivolumab_plus_ipilimumab cabozantinib_plus_nivolumab nivolumab_monotherapy cannot be in one permutation
Permutations before applying rule: 400
Permutations after applying rule : 400
[1] "TKIs"
applying rule TKIs : sunitinib pazopanib tivozanib cannot be in one permutation
Permutations before applying rule: 400
Permutations after applying rule : 202
[1] "lenvatinib_plus_everolimus"
applying rule: lenvatinib_plus_everolimus can only be after ONE of avelumab_plus_axitinib axitinib cabozantinib cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 202
Permutations after applying rule : 182
[1] "cabozantinib"
applying rule: cabozantinib as 2L+ only allowed after avelumab_plus_axitinib axitinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 182
Permutations after applying rule : 182
[1] "pazopanib"
applying rule: pazopanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 182
Permutations after applying rule : 172
[1] "sunitinib"
applying rule: sunitinib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 172
Permutations after applying rule : 162
[1] "tivozanib"
applying rule: tivozanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 162
Permutations after applying rule : 152
Applying sequence restrictions to population 2
Dropping drugs not allowed for this population.
applying rule: avelumab_plus_axitinib axitinib cabozantinib everolimus nivolumab_plus_ipilimumab lenvatinib_plus_everolimus cabozantinib_plus_nivolumab nivolumab_monotherapy pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib are only allowed treatments.
Permutations before applying rule: 26404
Permutations after applying rule : 13344
applying rule: drug line restrictions.
Permutations before applying rule: 13344
Permutations after applying rule : 1036
[1] "axitinib"
applying rule. axitinib is only allowed after avelumab_plus_axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 1036
Permutations after applying rule : 1017
[1] "everolimus"
applying rule. everolimus is only allowed after avelumab_plus_axitinib axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 1017
Permutations after applying rule : 1004
[1] "lenvatinib_plus_everolimus"
applying rule. lenvatinib_plus_everolimus is not allowed immediately after nivolumab_plus_ipilimumab
Permutations before applying rule: 1004
Permutations after applying rule : 984
[1] "axitinib"
applying rule axitinib : avelumab_plus_axitinib axitinib cannot be in one permutation
Permutations before applying rule: 984
Permutations after applying rule : 915
[1] "cabozantinib"
applying rule cabozantinib : cabozantinib cabozantinib_plus_nivolumab cannot be in one permutation
Permutations before applying rule: 915
Permutations after applying rule : 876
[1] "everolimus"
applying rule everolimus : lenvatinib_plus_everolimus everolimus cannot be in one permutation
Permutations before applying rule: 876
Permutations after applying rule : 773
[1] "io"
applying rule io : avelumab_plus_axitinib nivolumab_plus_ipilimumab cabozantinib_plus_nivolumab nivolumab_monotherapy lenvatinib_plus_pembrolizumab cannot be in one permutation
Permutations before applying rule: 773
Permutations after applying rule : 657
[1] "lenvatinib_plus_everolimus"
applying rule lenvatinib_plus_everolimus : lenvatinib_plus_everolimus lenvatinib_plus_pembrolizumab cannot be in one permutation
Permutations before applying rule: 657
Permutations after applying rule : 638
[1] "nivolumab"
applying rule nivolumab : nivolumab_plus_ipilimumab cabozantinib_plus_nivolumab nivolumab_monotherapy cannot be in one permutation
Permutations before applying rule: 638
Permutations after applying rule : 638
[1] "TKIs"
applying rule TKIs : sunitinib pazopanib tivozanib cannot be in one permutation
Permutations before applying rule: 638
Permutations after applying rule : 386
[1] "lenvatinib_plus_everolimus"
applying rule: lenvatinib_plus_everolimus can only be after ONE of avelumab_plus_axitinib axitinib cabozantinib cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 386
Permutations after applying rule : 359
[1] "cabozantinib"
applying rule: cabozantinib as 2L+ only allowed after avelumab_plus_axitinib axitinib nivolumab_plus_ipilimumab lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 359
Permutations after applying rule : 359
[1] "pazopanib"
applying rule: pazopanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 359
Permutations after applying rule : 339
[1] "sunitinib"
applying rule: sunitinib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 339
Permutations after applying rule : 319
[1] "tivozanib"
applying rule: tivozanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 319
Permutations after applying rule : 299
Applying sequence restrictions to population 3
Dropping drugs not allowed for this population.
applying rule: axitinib cabozantinib everolimus lenvatinib_plus_everolimus nivolumab_monotherapy pazopanib sunitinib tivozanib are only allowed treatments.
Permutations before applying rule: 26404
Permutations after applying rule : 2080
applying rule: drug line restrictions.
Permutations before applying rule: 2080
Permutations after applying rule : 330
[1] "axitinib"
applying rule. axitinib is only allowed after avelumab_plus_axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 330
Permutations after applying rule : 330
[1] "everolimus"
applying rule. everolimus is only allowed after avelumab_plus_axitinib axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 330
Permutations after applying rule : 330
[1] "lenvatinib_plus_everolimus"
applying rule. lenvatinib_plus_everolimus is not allowed immediately after nivolumab_plus_ipilimumab
Permutations before applying rule: 330
Permutations after applying rule : 330
[1] "axitinib"
applying rule axitinib : avelumab_plus_axitinib axitinib cannot be in one permutation
Permutations before applying rule: 330
Permutations after applying rule : 330
[1] "everolimus"
applying rule everolimus : lenvatinib_plus_everolimus everolimus cannot be in one permutation
Permutations before applying rule: 330
Permutations after applying rule : 288
[1] "lenvatinib_plus_everolimus"
applying rule lenvatinib_plus_everolimus : lenvatinib_plus_everolimus lenvatinib_plus_pembrolizumab cannot be in one permutation
Permutations before applying rule: 288
Permutations after applying rule : 288
[1] "TKIs"
applying rule TKIs : sunitinib pazopanib tivozanib cannot be in one permutation
Permutations before applying rule: 288
Permutations after applying rule : 120
[1] "lenvatinib_plus_everolimus"
applying rule: lenvatinib_plus_everolimus can only be after ONE of axitinib cabozantinib pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 120
Permutations after applying rule : 111
[1] "cabozantinib"
applying rule: cabozantinib as 2L+ only allowed after avelumab_plus_axitinib axitinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 111
Permutations after applying rule : 111
[1] "pazopanib"
applying rule: pazopanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 111
Permutations after applying rule : 111
[1] "sunitinib"
applying rule: sunitinib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 111
Permutations after applying rule : 111
[1] "tivozanib"
applying rule: tivozanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 111
Permutations after applying rule : 111
Applying sequence restrictions to population 4
Dropping drugs not allowed for this population.
applying rule: axitinib cabozantinib everolimus lenvatinib_plus_everolimus nivolumab_monotherapy pazopanib sunitinib tivozanib are only allowed treatments.
Permutations before applying rule: 26404
Permutations after applying rule : 2080
applying rule: drug line restrictions.
Permutations before applying rule: 2080
Permutations after applying rule : 440
[1] "axitinib"
applying rule. axitinib is only allowed after avelumab_plus_axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 440
Permutations after applying rule : 440
[1] "everolimus"
applying rule. everolimus is only allowed after avelumab_plus_axitinib axitinib cabozantinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 440
Permutations after applying rule : 440
[1] "lenvatinib_plus_everolimus"
applying rule. lenvatinib_plus_everolimus is not allowed immediately after nivolumab_plus_ipilimumab
Permutations before applying rule: 440
Permutations after applying rule : 440
[1] "axitinib"
applying rule axitinib : avelumab_plus_axitinib axitinib cannot be in one permutation
Permutations before applying rule: 440
Permutations after applying rule : 440
[1] "everolimus"
applying rule everolimus : lenvatinib_plus_everolimus everolimus cannot be in one permutation
Permutations before applying rule: 440
Permutations after applying rule : 384
[1] "lenvatinib_plus_everolimus"
applying rule lenvatinib_plus_everolimus : lenvatinib_plus_everolimus lenvatinib_plus_pembrolizumab cannot be in one permutation
Permutations before applying rule: 384
Permutations after applying rule : 384
[1] "TKIs"
applying rule TKIs : sunitinib pazopanib tivozanib cannot be in one permutation
Permutations before applying rule: 384
Permutations after applying rule : 198
[1] "lenvatinib_plus_everolimus"
applying rule: lenvatinib_plus_everolimus can only be after ONE of axitinib cabozantinib pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 198
Permutations after applying rule : 182
[1] "cabozantinib"
applying rule: cabozantinib as 2L+ only allowed after avelumab_plus_axitinib axitinib lenvatinib_plus_everolimus cabozantinib_plus_nivolumab pazopanib lenvatinib_plus_pembrolizumab sunitinib tivozanib
Permutations before applying rule: 182
Permutations after applying rule : 182
[1] "pazopanib"
applying rule: pazopanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 182
Permutations after applying rule : 182
[1] "sunitinib"
applying rule: sunitinib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 182
Permutations after applying rule : 182
[1] "tivozanib"
applying rule: tivozanib as 2L+ only allowed immediately after avelumab_plus_axitinib lenvatinib_plus_pembrolizumab cabozantinib_plus_nivolumab nivolumab_plus_ipilimumab
Permutations before applying rule: 182
Permutations after applying rule : 182