5 Reproduction
5.1 Studies
Shoaib and Ramamohan (2021): Uses python (salabim) to model primary health centres (PHCs) in India. The model has four patient types: outpatients, inpatients, childbirth cases and antenatal care patients. Four model configurations are developed based on observed PHC practices or government-mandated operational guidelines. The paper explores different operational patterns for scenarios where very high utilisation was observed, to explore what might help reduce utilisation of these resources. Note: The article was as Shoaib and Ramamohan (2022), but we used the green open access pre-print Shoaib and Ramamohan (2021). Link to reproduction.
Huang et al. (2019): Uses R (simmer) to model an endovascular clot retrieval (ECR) service. ECR is a treatment for acute ischaemic stroke. The model includes the stroke pathway, as well as three other pathways that share resources with the stroke pathway: an elective non-stroke interventional neuroradiology pathway, an emergency interventional radiology pathway, and an elective interventional radiology pathway. The paper explores waiting times and resource utilisation - particularly focussing on the biplane angiographic suite (angioINR). A few scenarios are tried to help examine why the wait times are so high for the angioINR. Link to reproduction.
Lim et al. (2020): Uses python (NumPy and pandas) to model the transmission of COVID-19 in a laboratory. It examines the proportion of staff infected in scenarios varying the: number of shifts per day; number of staff per shift; overall staff pool; shift patterns; secondary attack rate of the virus; introduction of protective measures (social distancing and personal protective equipment). Link to reproduction.
Kim et al. (2021): Adapts a previously developed R (Rcpp, expm, msm, foreach, iterators, doParallel) model for abdominal aortic aneurysm (AAA) screening of men in England. The model is adapted/used to explore different approaches to resuming screening and surgical repair for AAA, as these services were paused or substantially reduced during COVID-19 due to concerns about virus transmission. Link to reproduction.
Anagnostou et al. (2022): This paper includes two models - we have focussed just on the dynamiC Hospital wARd Management (CHARM) model. CHARM uses python (SimPy) to model intensive care units (ICU) in the COVID-19 pandemic (as well as subsequent stays in a recovery bed). It includes three types of admission to the ICU (emergency, elective or COVID-19). COVID-19 patients are kept seperate, and if they run out of capacity due to a surge in COVID-19 admissions, additional capacity can be pooled from the elective and emergency capacity. Link to reproduction.
Johnson et al. (2021): This study uses a previously validated discrete-event simulation model, EPIC: Evaluation Platform in chronic obstructive pulmonary disease (COPD). The model is written in C++ with an R interface, using R scripts for execution. The model is adapted to evaluate the cost-effectiveness of 16 COPD case detection strategies in primary care, comparing costs, quality-adjusted life years (QALYs), incremental cost-effectiveness ratios (ICER), and incremental net monetary benefits (INMB) across scenarios. Sensitivity analyses are also conducted. Link to reproduction.
Hernandez et al. (2015): This study models Points-of-Dispensing (PODs) in New York City. These are sites set up during a public health emergency to dispense counter-measures. The authors use evolutionary algorithms combined with discrete-event simulation to explore optimal staff numbers with regards to resource use, wait time and throughput. They use python for most of the analysis (with SimPy for the simulation component), but R to produce the plots and tables for the paper. Link to reproduction.
Wood et al. (2021): This study uses discrete-event simulation (R (base R, dplyr, tidyr)) to explore the deaths and life years lost under different triage strategies for an intensive care unit, relative to a baseline strategy. The unit is modelled with 20 beds (varied from 10 to 200 in sensitivity analyses). Three different triage strategies are explored, under three different projected demand trajectories. Link to reproduction.
5.2 Scope and reproduction
| Study | Scope | Success | Time |
|---|---|---|---|
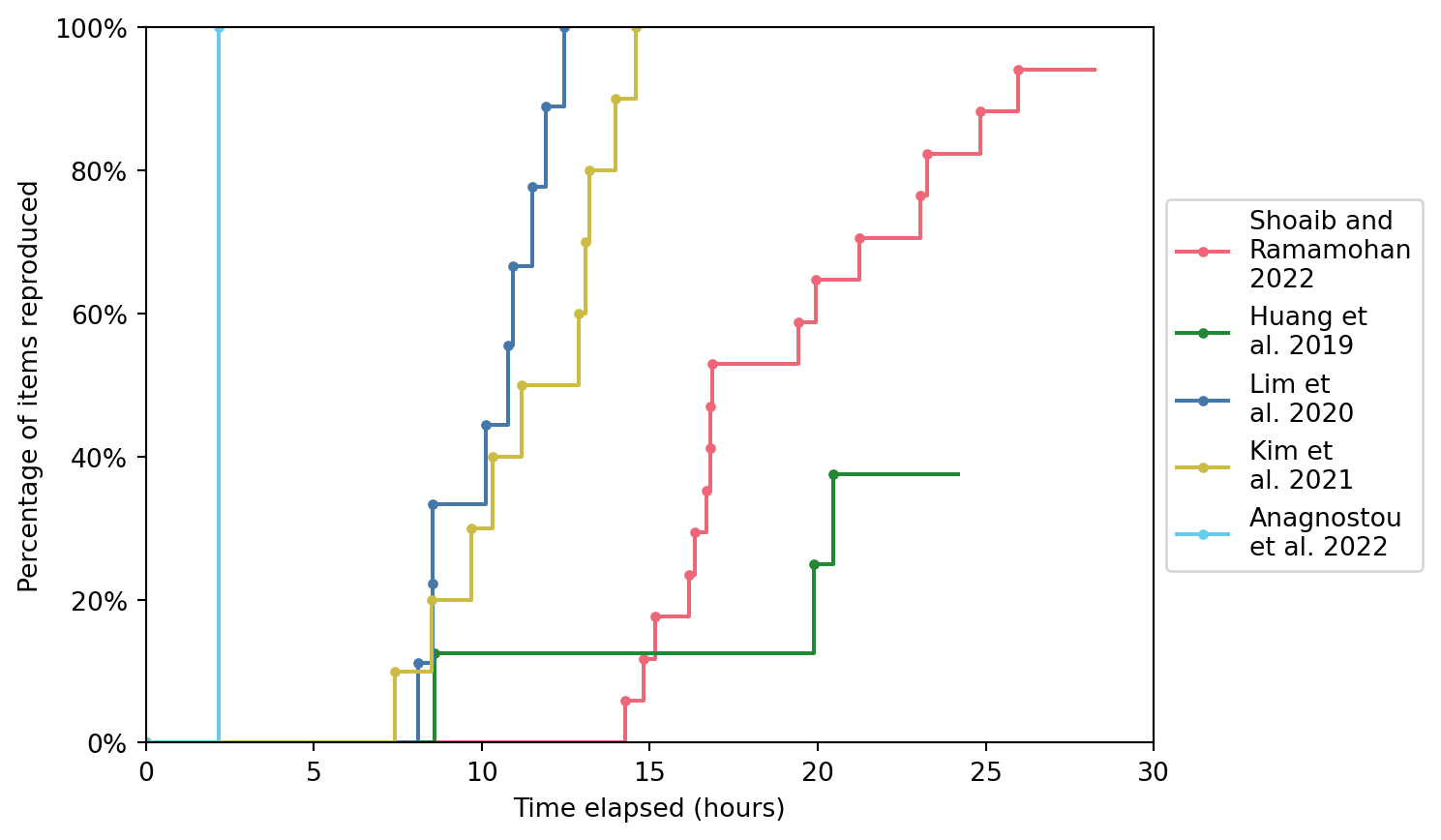
| Shoaib and Ramamohan 2022 | 17 items: • 1 table • 9 figures • 7 in-text results |
16 out of 17 (94.1%) | 28h 14m |
| Huang et al. 2019 | 8 items: • 5 figures • 3 in-text results |
3 out of 8 (37.5%) | 24h 10m |
| Lim et al. 2020 | 9 items: • 5 tables • 4 figures |
9 out of 9 (100%) | 12h 27m |
| Kim et al. 2021 | 10 items: • 3 tables • 6 figures • 1 in-text result |
10 out of 10 (100%) | 14h 42m |
| Anagnostou et al. 2022 | 1 item: • 1 figure |
1 out of 1 (100%) | 2h 11m |
| Johnson et al. 2021 | 5 items: • 1 table • 4 figures |
4 out of 5 (80%) | 19h 49m |
| Hernandez et al. 2015 | 8 items: • 6 figures • 2 tables |
1 out of 8 (12.5%) | 17h 56m |
| Wood et al. 2021 | 5 items: • 4 figures • 1 table |
5 out of 5 (100%) | 3h 50m |
5.3 Time to complete reproductions
Non-interactive figure:
Version of non-interactive figure used in article:

Interactive figure:
5.4 Model run times
For reference, the run times for the models are detailed below. It’s worth being aware that these are not compared on a “level playing field” as they were run on different machines, and with/without parallel processing or parallel terminals.
| Study | Machine specs | Model run time | Additional details |
|---|---|---|---|
| Shoaib and Ramamohan (2021) | Intel Core i7-12700H with 32GB RAM running Ubuntu 22.04.4 Linux | 22 minutes 26 seconds | This is based on the combined runtime of the notebooks which use 10 replications (rather than 100) and parallel processing, but with each notebook run seperately. |
| Huang et al. (2019) | Intel Core i7-12700H with 32GB RAM running Ubuntu 22.04.4 Linux | 29 minutes 10 seconds | Combined time from notebooks run seperately. |
| Lim et al. (2020) | Intel Core i7-12700H with 32GB RAM running Ubuntu 22.04.4 Linux | 49 minutes and 17 seconds | |
| Kim et al. (2021) | Intel Core i9-13900K with 81GB RAM running Pop!_OS 22.04 Linux | 6 hours 53 minutes | We reduced the number of patients in the simulation from 10 million to 1 million, to improve run times. Note, you can expect the runtime to be notably longer on machines with lower specs than this. For example, I ran surveillance scenario 0 on an Intel Core i7-12700H with 32GB RAM running Ubuntu 22.04.4 Linux. The runtime increased from 4 minutes 28 seconds up to 21 minutes 59 seconds. |
| Anagnostou et al. (2022) | Intel Core i7-12700H with 32GB RAM running Ubuntu 22.04.4 Linux | Only a few seconds. | |
| Johnson et al. (2021) | Intel Core i9-13900K with 81GB RAM running Pop!_OS 22.04 Linux | At least 1 day 11 hours | It was run from the command line via multiple terminals simultaneously - hence, the exact times are impacted by the number being ran at once. The base case scenarios were ran at the same time, and took an overall total time of 20 hours 40 minutes. The sensitivity analysis scenario were all ran at the same time, and took an overall total of 1 day 10 hours 57 minutes. Four files (scenarios 2 and 3) had to be re-run seperately after fixing a mistake, and when just those were run, the total was 21 hours 26 minutes (quicker than when they were run as part of the full set of scenarios, when they took up to 1.3 days). Based on this, if you were to run all scenarios at once in parallel in seperate terminals, you could expect a run time of at least 1 day 11 hours, but would be higher than that (since the more run at once, the lower it takes, as you can see from the scenario 2 and 3 times above). |
| Hernandez et al. (2015) | Intel Core i9-13900K with 81GB RAM running Pop!_OS 22.04 Linux | 9 hours 16 minutes | This involved running the models within each experiment in parallel, but each of the experiment files seperately. If these are run at the same time (which I could do without issue), then you will be able to run them all within 4 hours 28 minutes (the longest experiment) (or a little longer, due to slowing speeds from running at once). Also, this has excluded one of the variants for Experiment 3, which I did not run as it had a very long run time (quoted to be 27 hours in the article) and as, regardless, I had not managed to reproduce the other sub-plots in the figure for that experiment. |
| Wood et al. (2021) | Intel Core i9-13900K with 81GB RAM running Pop!_OS 22.04 Linux | 48 hours 25 minutes | It ran in a single loop, so required the computer to remain on for that time. This time includes all scenarios - but, for just the base scenario, the run time was 2 hours 3 minutes. |