3 Methods
For work package 1, the methods were described in our protocol, Heather et al. (2024).
Heather, Amy, Thomas Monks, Alison Harper, Navonil Mustafee, and Andrew Mayne. 2024. “Protocol for Assessing the Computational Reproducibility of Discrete-Event Simulation Models on STARS,” June. https://zenodo.org/records/12179846.
These are summarised below, and modifications from the protocol are also described.
3.1 Protocol summary
For this work, eight published healthcare DES models were selected. These were models with publicly available code under an open licence (either already available or add on request from the STARS team). For each model, the follow stages of work were conducted:
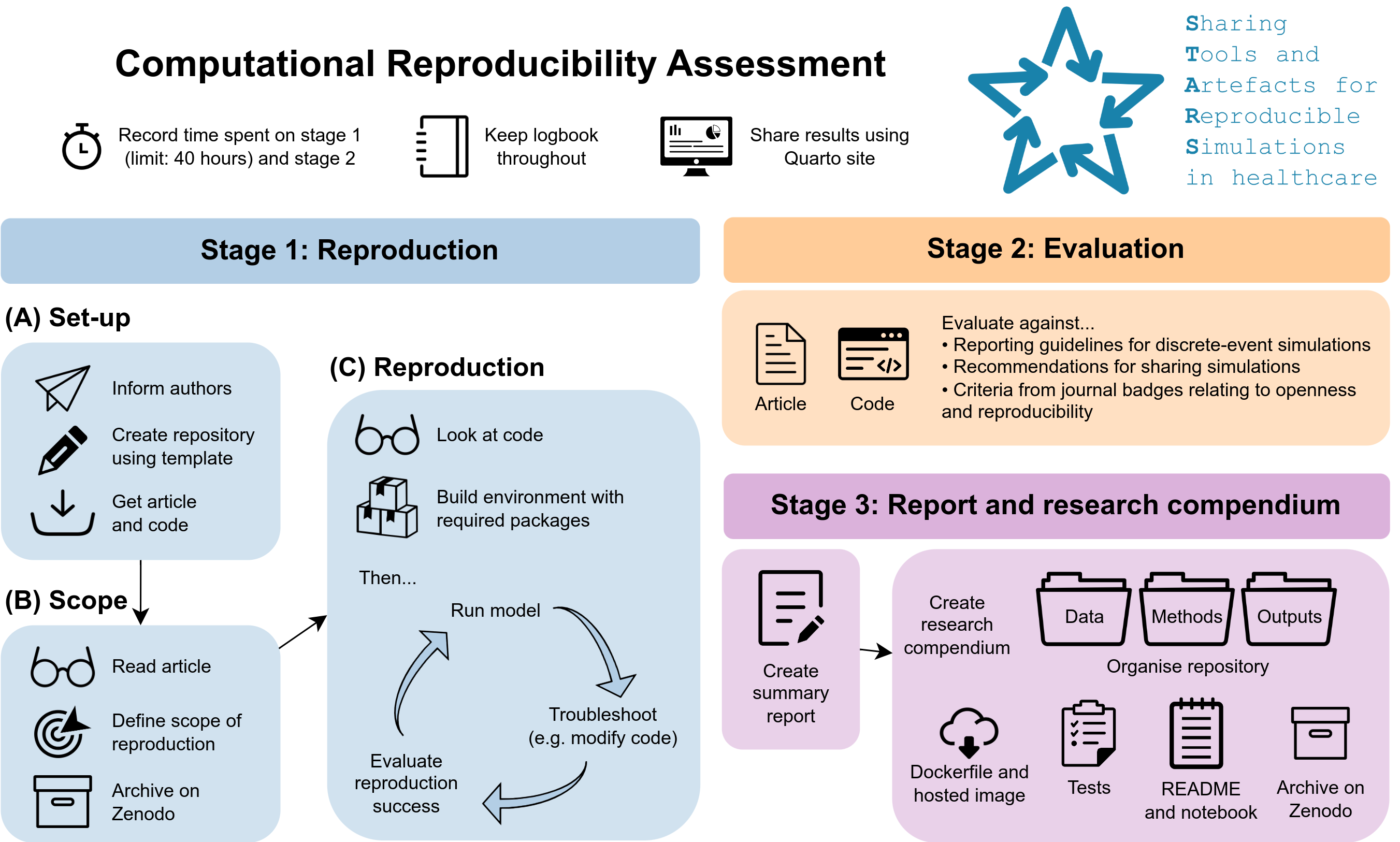
Stage 1: Reproduction - assessing the computational reproducibility of each study
- Informed authors about study and, if not available, asked if they would be happy to add an open licence to their code
- Set up repository for reproduction with the article and code
- Read the article and defined the scope of the reproduction, archiving the scope (and repository) on Zenodo before proceeding
- Looked over the model code, created a suitable environment with the software and packages required, and then ran the model. For each study, with any issues faced in running the model, troubleshooting was performed such as modifying or writing code. If troubleshooting was exhaused and there were still issues or discrepancies in the results, the original study authors were informed and provided the opportunity to advice on the reason for this (although with no pressure or requirement to do so)
- For each item in the scope, a decision was made as to whether this had been successfully reproduced or not. This was a subjective decision which allowed some expected deviation due to model stochasticity (for example, lack of seed control).
- This is timed (including time to produce each item in the scope), and limited to a maximum of 40 horus
Stage 2: Evaluation - evaluating the publication, code and associated artefacts against sharing and reporting standards
- The publication was evaluated against reporting guidelines for DES:
- Monks et al. (2019) - STRESS-DES: Strengthening The Reporting of Empirical Simulation Studies (Discrete-Event Simulation) (Version 1.0).
- Zhang, Lhachimi, and Rogowski (2020) - The generic reporting checklist for healthcare-related discrete event simulation studies derived from the the International Society for Pharmacoeconomics and Outcomes Research Society for Medical Decision Making (ISPOR-SDM) Modeling Good Research Practices Task Force reports.
- The model code and associated artefacts (e.g. the GitHub repository shared by the authors) was evaluated against:
- The criteria of badges related to reproducibility from various organisations and journals - namely:
- National Information Standards Organisation (NISO)(NISO Reproducibility Badging and Definitions Working Group (2021))
- Association for Computing Machinery (ACM) (Association for Computing Machinery (ACM) (2020))
- Center for Open Science (COS) (Blohowiak et al. (2023))
- Institute of Electrical and Electronics Engineers (IEEE) (Institute of Electrical and Electronics Engineers (IEEE) (2024))
- Psychological Science (Hardwicke and Vazire (2024) and Association for Psychological Science (APS) (2024))
- Recommendations from the pilot STARS framework for the sharing of code and associated materials from discrete-event simulation models (Monks, Harper, and Mustafee (2024)).
- The criteria of badges related to reproducibility from various organisations and journals - namely:
- This is timed
Stage 3: Report and research compendium - summary report and organised repository
- Wrote a report summarising the computational reproducibility assessment and evaluation
- Restructed the reposuitory into a “research compendium”, which essentially consisted of organising the repository to ensure it is easy and clear for someone else to re-run. Steps included:
- Adding run times to the model notebooks
- Write a README for the reproduction folder
- Moving data, methods and outputs into seperate folders
- Creating tests which check if a user can get the same results from the model as we did during the reproduction
- A Dockerfile and Docker image published on the GitHub container registry
- A second researcher from the STARS team then attempted to use the repository and confirm whether they were able to reproduce the results of the first researcher
- Finally, the repository was archived on Zenodo, and the authors were informed.
For each study, a quarto site was produced which shared the results from the reproduction and evaluation and the summary report. Throughout the work, a detailed logbook was kept to keep track of timings and to record work on each stage, such as detailing troubleshooting steps during the reproduction, or uncertainities discussed with another STARS team member during the evaluation.
Summary diagram
This process is captured in the diagram below:
3.2 Inspiration
As cited in Heather et al. (2024), the protocol was informed by several prior studies assessing computational reproducibility:
- Krafczyk et al. (2021): Assessed the reproducibility of seven articles published in the Journal of Computational Physics.
- B. D. K. Wood, Müller, and Brown (2018) and B. Wood et al. (2018): Assessed the reproducibility of 109 published impact evaluations in low- and middle-income countries. Conducted in association with the replication programme of the International Initiative for Impact Evaluation (3ie).
- Schwander et al. (2021): Assessed the reproducibility of four health economic obesity models.
- Laurinavichyute, Yadav, and Vasishth (2022): Assessed the reproducibility of 118 articles published in the Journal of Memory and Language.
- Konkol, Kray, and Pfeiffer (2019): Assessed the reproducibility of 41 geoscientific articles from Copernicus Publications and the Journal of Statistical Software.
It was also informed by:
- Ayllón et al. (2021): Article with recommendations on keeping modelling notebooks to support completion of TRACE (TRAnsparent and Comprehensive model Evaluation) documents.
- Berkeley Initiative for Transparency in the Social Sciences (2022): Guidelines on conducting reproductions of published social science research.
- McManus, Turner, and Sach (2019): Article proposing several possible definitions for success in reproducing or replicating models in health economics.
- Marwick, Boettiger, and Mullen (2018): Article recommending how to structure data analytical work as research compendiums using the R package structure.
3.3 Modifications from the protocol
There were two main changes from the protocol…
| Modification | Description and reason for the change |
|---|---|
| Expanding from 6 to 8 studies | Whilst partway through the 6th study, we reflected on what we had found so far, and felt it would be beneficial to do a few more studies. The primary motivation for this was to try and include a selection of studies that reflect the range of studies in the literature. So far, we had a few studies where the model code/run times were very large or very small, and we felt it beneficial to try and include some more “medium-sized” studies, as these are more typical of the literature. We also thought it would be good to include an older study (for example, about 10 years ago, if we can find one), as all those included so far were within the last few years, but working with more outdated code will be/can be a problem people encounter. |
| Using the latest software packages | In the protocol, I had planned that - if no versions were provided we select a version of the software and each package that is closest to but still prior to the date of the code archive or paper publication. I kept to this for the Python models (easily set using a conda/mamba environment). However, I had great difficulties attempting to do this in R, and could not successfully backdate both. As such, I used the latest versions of R and each package for those studies |
There were also some very minor changes, which are explained below…
| Modification | Description and reason for the change |
|---|---|
| Using percentage difference in results to help decided reproduction success | I did explore this, but I ultimately found it very unhelpful, as the percentage difference could be greatly impacted by scale (for example, 0.1 vs 0.2 would appear much greater than 3 vs 4, but the actual meaning of these differences might be similar (e.g. both might be considered a small difference) depending on the scale used and what is being compared - whilst in another context with a different scale, 0.1 vs 0.2 might actually reflect a huge difference!). |
| Moving onto evaluation stage before receive author response regarding reproduction discrepancy or before getting consensus on reproduction | In the protocol, we required that authors are contacted if there are any remaining difficulties in running the code or items in the scope that were not reproduced. The authors were given a total of four weeks to respond if they chose to. We had implied that we must wait for this time to pass before continuing to the evaluation stage (since the three stages were presented as being completed one after another). The rationale for this was that the timings for the reproduction would be influenced by whether the evaluation had been completed or not, and vice versa. However, given the many possible influences on the timings, this was considered negligble. We also required consensus on whether items had been succesfully reproduced or not before moving on. In some cases, this was not done, as other team members were not available (e.g. busy or on annual leave) and so could not yet give a second opinion, and so I progressed with the evaluation and got consensus on reproductions afterwards, once they were available. |
| Organisation of the repository for the research compendium | In the protocol, we had planned that seperate folders were created for data, methods and outputs. This was generally followed but, if an alternative structure seemed more suitable. For example, if the original study already divided items well, but perhaps with different folder names or with multiple scripts folders or so on, we might have used that original structure, as it still served the purpose of being clear and easy to re-run, whilst reducing the number of differences compared with the original study. |
| Retrospective archiving | For Johnson et al. 2021, I forgot to create a release to archive this repository after defining scope and before proceeding with reproduction. However, I was easily able to resolve this by setting the release to a prior commit, choosing the commit from that timepoint (between scope and reproduction), so it was as if it had been done at the time |
| Identification of papers | Although 7 papers were identified from the review by Monks and Harper (2023), 1 was identified from additional searches (informal, not systematic) - this was so we could find an older paper. |
| Reflections | I effectivelly created an “additional” stage, which was to create a page of reflections on the reproduction (barriers and faciliators). This was not formally part of the original protocol, but implicit. |
3.4 Timings
As per the protocol, the reproduction and evaluation stages were timed. Although this was conducted carefully and thoroughly, it will not be perfect, and we recognise some of the potential sources of variation in timings between studies, such as:
- Whether we consistently included amendments to the quarto site and repository and time spent on GitHub commits etc. within the timings for the reproduction.
- For the first R study (Huang et al. (2019)), I initially tried to create an environment with R and package versions prior to the article publication date, although had great difficulties with this and ended up using the latest versions. This contributed to the set-up time during this reproduction, but on later R models (Kim et al. (2021) and Johnson et al. (2021)), based on that experience, I did not attempt to backdate them when getting started.
- Any estimated times (for example, if I were partway through working but someone in the office came to talk to me and I forgot to note the time of that, I might estimate if that were about five or ten minutes of conversation, and set the time accordingly).
- Timings from consensus discussions regarding uncertainities in the evaluation or reproduction (as these might be longer if done in person rather than over email - or vice versa - and I sometimes spent longer on sorting/tidying these for some studies than others, which I would have included in the time)
- Whether subjectively feel that need to add random seeds during reproduction stage, if results vary considerably between each run, and so a certain seed could get a much more similar result than another
At an estimate, this uncertainty between study timings would lead me to conclude that the timings are approximately correct, give or take up to about four hours. However, this is just an estimate, and it is worth noting that Krafczyk et al. (2021), who also conducted computational reproducibility assessments in a different context, estimated that human error introduced a maximum of 8 hours ambiguity in the timings, due to the “non-precise nature of starting and stopping the watch consistently”.