Take-home message:
There are simple steps researchers can take to improve the reproducibility of their healthcare DES models. These include:
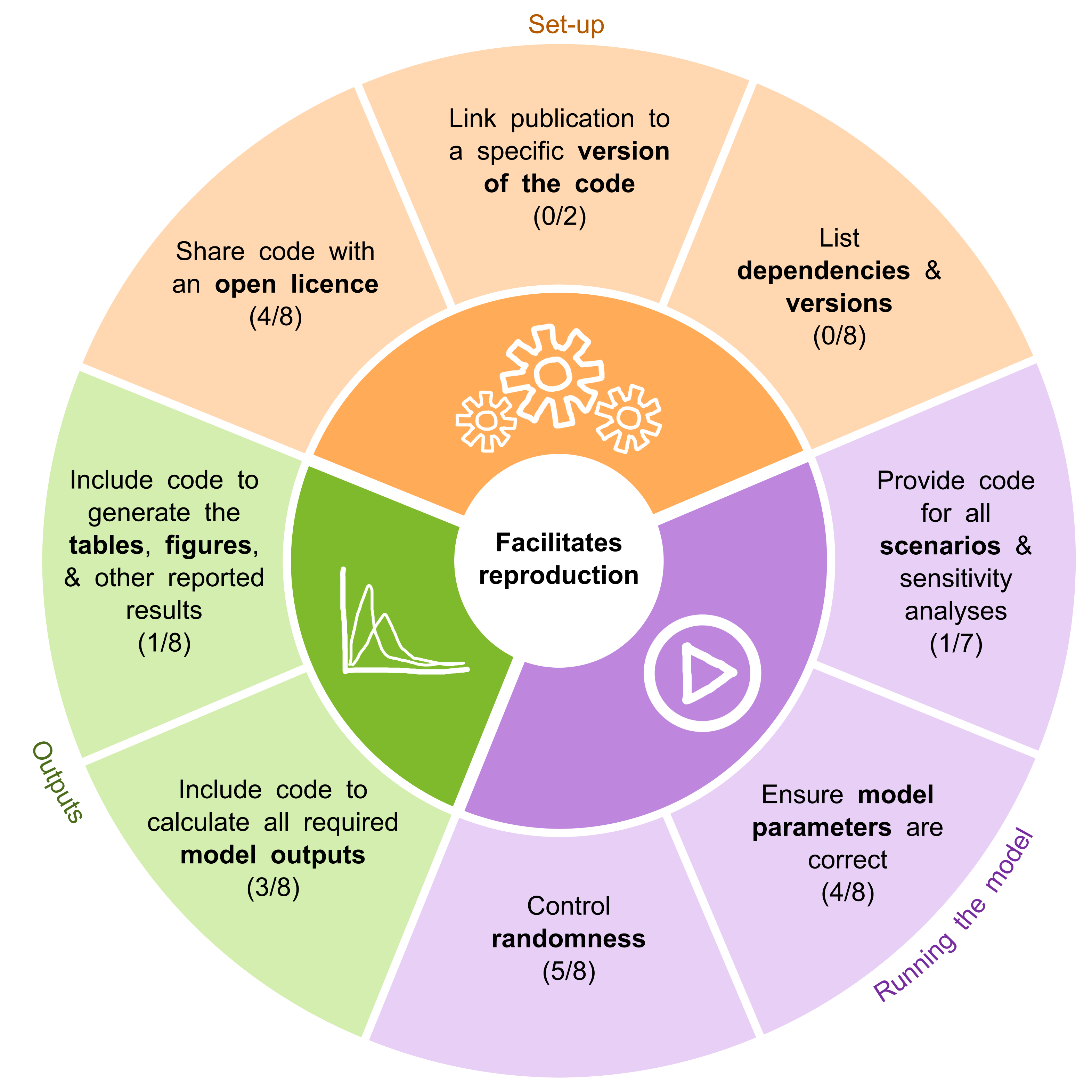
- Sharing code with an open licence.
- Ensuring model parameters are correct.
- Including code to calculate all required model outputs.
- Providing code for all scenarios and sensitivity analyses.
- Including code to generate the tables, figures, and other reported results.
For more suggestions, see the STARS reproducibility recommendations below.
Summary
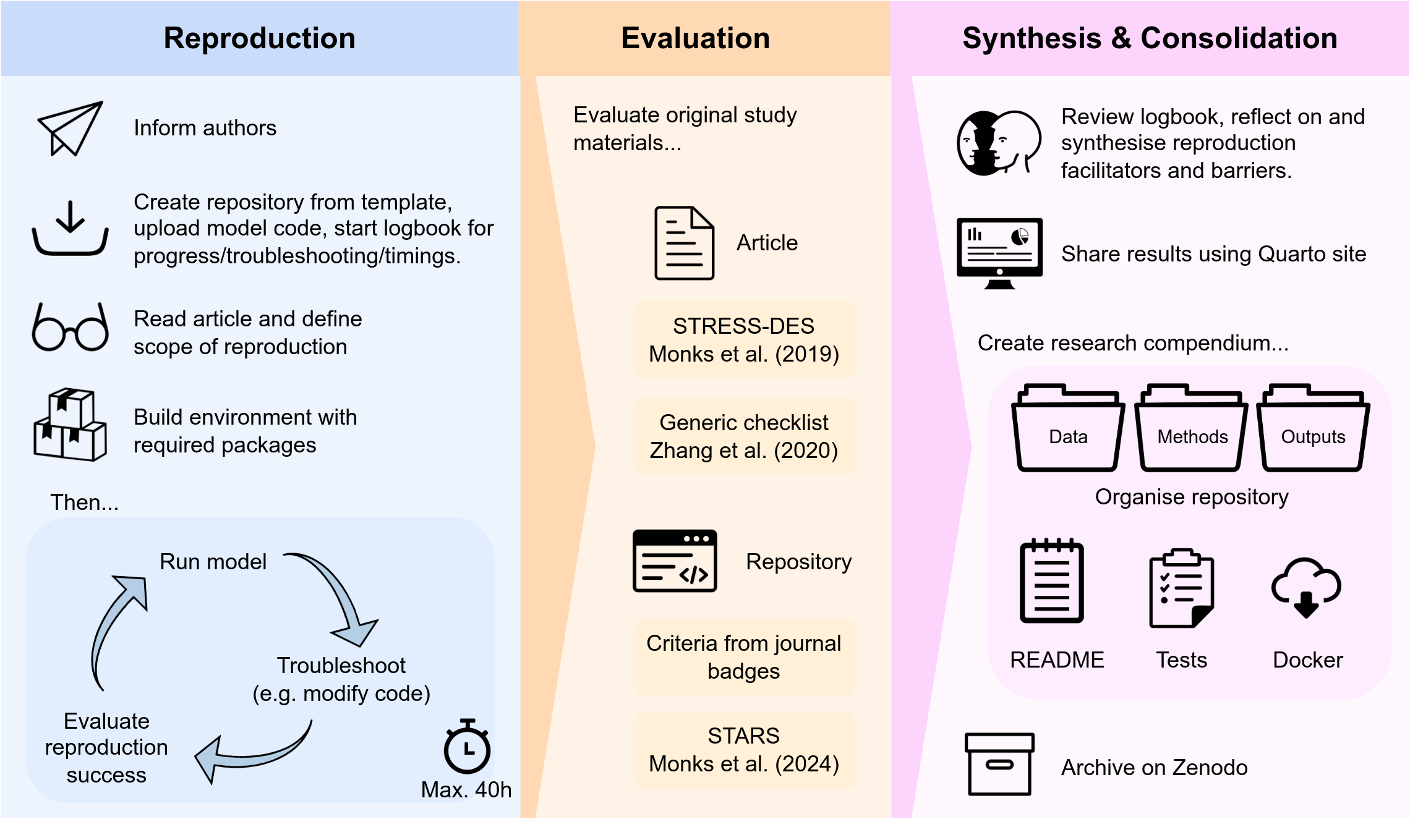
This work assessed the computational reproducibility of eight published healthcare DES models implemented in Python or R. A detailed study protocol was first developed, informed by existing reproducibility studies and pilot work. The workflow for assessing each study is summarised in the figure below.
The eight models were selected to ensure diversity across a range of factors including the health focus (e.g., healthcare condition, specific system), geographical context, and model complexity
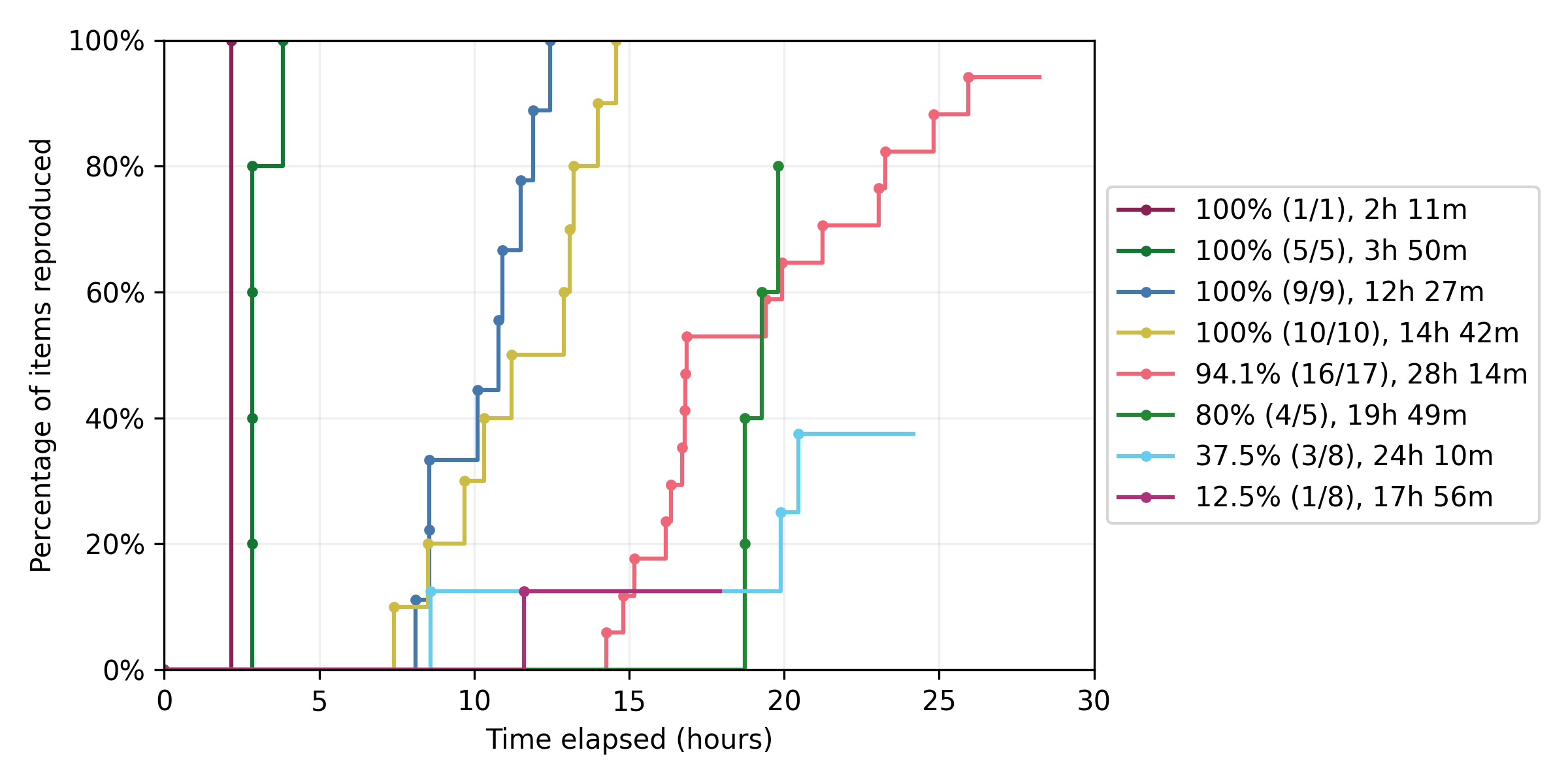
Reproducing results required up to 28 hours of troubleshooting per model. Four models were judged to be fully reproduced, while four were partially reproduced - between 12.5% and 94.1% of reported outcomes.
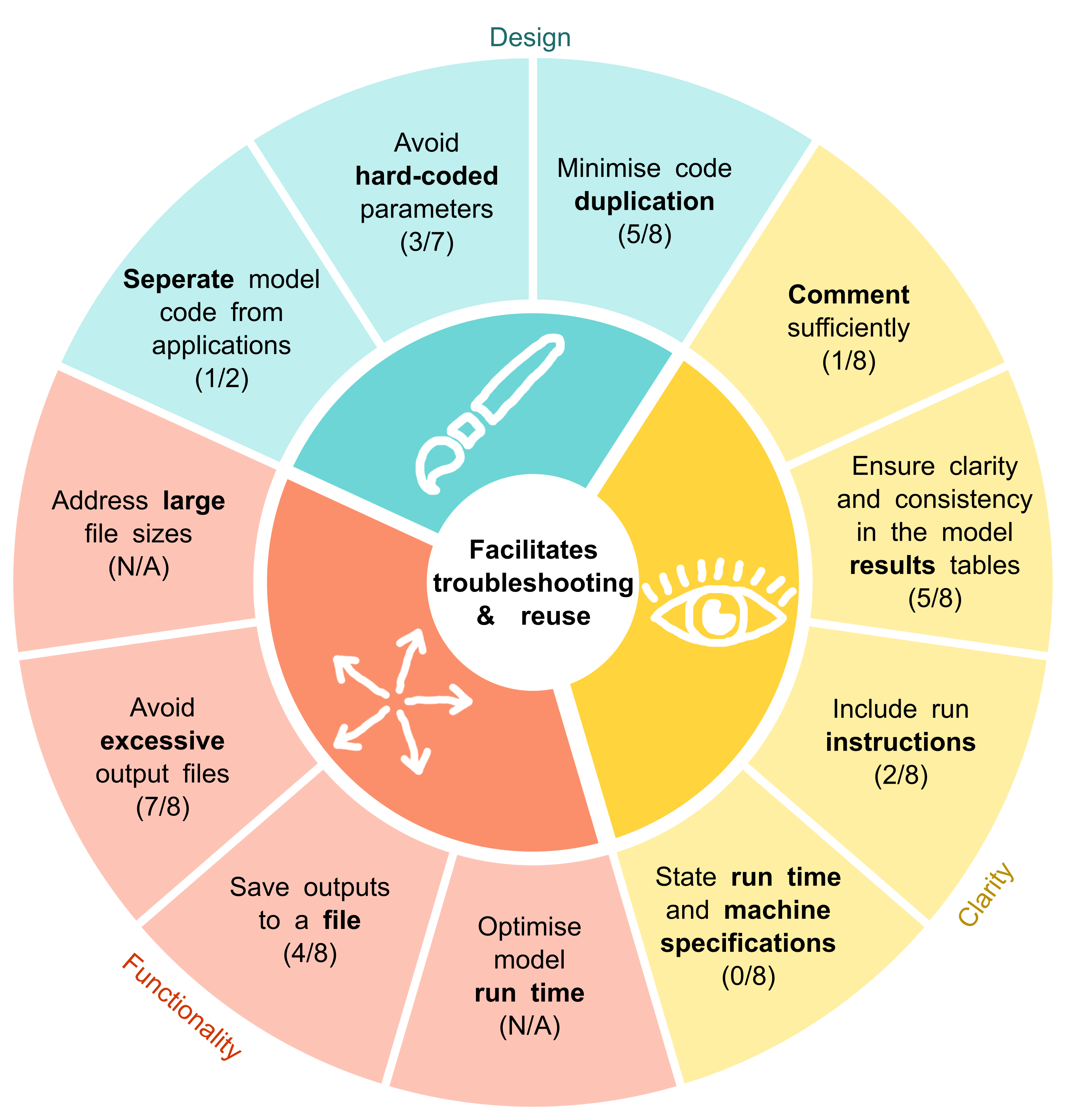
Based on the barriers and facilitators observed during these reproductions, we developed the STARS reproducibility recommendations. These are presented in the figures below, divided into categories: recommendations that specifically support reproducibility, and those that were more relevant to troubleshooting models (and therefore also to reuse).
Websites and GitHub repositories
We have described this work in a publication in the Journal of Simulation.
The work is also documented in a dedicated Quarto summary website, which provides more fine-grained detail on the reproductions.
Each of the eight DES models has its own research compendium-style GitHub repository, with a corresponding website and archival record, as linked in the table below. Each repository was created from a template which we developed during pilot work. In pilot work, an example model from colleagues was used to test and refine the reproducibility protocol, resulting in the repository stars-reproduce-allen-2020.
| Reproduction study | Website | GitHub | Zenodo |
|---|---|---|---|
| Shoaib and Ramamohan 2022 | Website | stars-reproduce-shoaib-2022 | 10.1177/00375497211030931 |
| Huang et al. 2019 | Website | stars-reproduce-huang-2019 | 10.5281/zenodo.12657280 |
| Lim et al. 2020 | Website | stars-reproduce-lim-2020 | 10.5281/zenodo.12795365 |
| Kim et al. 2021 | Website | stars-reproduce-kim-2021 | 10.5281/zenodo.13121136 |
| Anagnostou et al. 2022 | Website | stars-reproduce-anagnostou-2022 | 10.5281/zenodo.13306159 |
| Johnson et al. 2021 | Website | stars-reproduce-johnson-2021 | 10.5281/zenodo.13832333 |
| Hernandez et al. 2015 | Website | stars-reproduce-hernandez-2015 | 10.5281/zenodo.13832260 |
| Wood et al. 2021 | Website | stars-reproduce-wood-2021 | 10.5281/zenodo.13881986 |
GW4 Open Research Prize - Improving Quality
This research was shortlisted for the “Improving Quality” prize, which is “for those able to demonstrate that the quality of their research has been enhanced through the adoption of open research practices in their work”. You can find out more about the prize and the other shortlisted entries and winners here.
Amy Heather presented this work at the GW4 Open Research Prize awards event. The slides were created using Quarto: see the slides and accompanying slides GitHub repository. A recording of the presentation is available:
Publications
This page was written by Amy Heather and reflects her interpretation of this work, which may not fully represent the views of all project authors or affiliated institutions.