SimPy: Treatment Centre#
Running the model
This version of the simulation model is in this notebook and can be executed online. You do not need to install anything on your computer. Move you mourse cursor over the rocket ship icon above and choose one of the two options: Binder (launches an external remote instance of Jupyter-Lab) or Thebe (code cells are excutable within the book).
See detailed instructions for more help.
1. Imports#
The model is provided with a conda virtual environment stars_treat_sim. Details are available in the Github repo.
import simpy
simpy.__version__
'4.1.1'
import numpy as np
import pandas as pd
import itertools
import math
import matplotlib.pyplot as plt
import fileinput
2. Constants and defaults for modelling as-is#
2.1 Distribution parameters#
# sign-in/triage parameters
DEFAULT_TRIAGE_MEAN = 3.0
# registration parameters
DEFAULT_REG_MEAN = 5.0
DEFAULT_REG_VAR= 2.0
# examination parameters
DEFAULT_EXAM_MEAN = 16.0
DEFAULT_EXAM_VAR = 3.0
DEFAULT_EXAM_MIN = 0.5
# trauma/stabilisation
DEFAULT_TRAUMA_MEAN = 90.0
# Trauma treatment
DEFAULT_TRAUMA_TREAT_MEAN = 30.0
DEFAULT_TRAUMA_TREAT_VAR = 4.0
# Non trauma treatment
DEFAULT_NON_TRAUMA_TREAT_MEAN = 13.3
DEFAULT_NON_TRAUMA_TREAT_VAR = 2.0
# prob patient requires treatment given trauma
DEFAULT_NON_TRAUMA_TREAT_P = 0.60
# proportion of patients triaged as trauma
DEFAULT_PROB_TRAUMA = 0.12
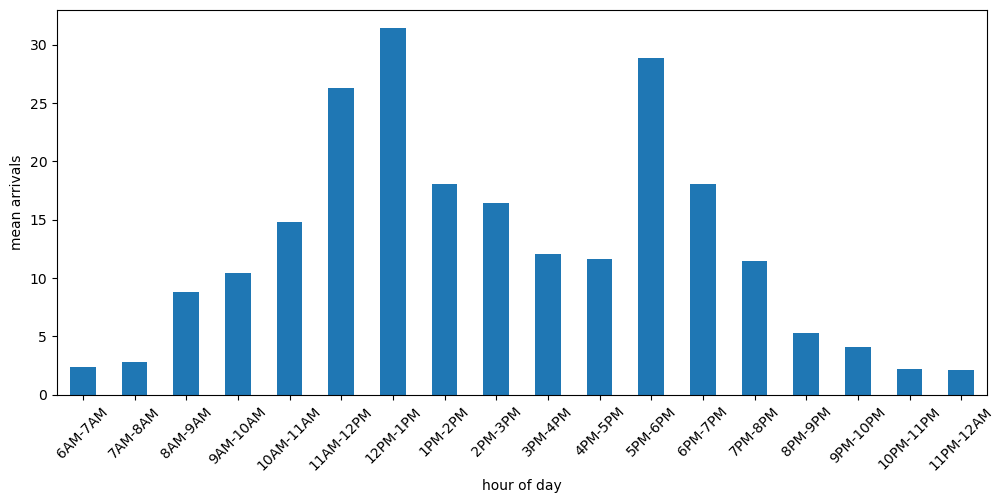
2.2 Time dependent arrival rates data#
The data for arrival rates varies between clinic opening at 6am and closure at 12am.
# data are held in the Github repo and loaded from there.
NSPP_PATH = 'https://raw.githubusercontent.com/TomMonks/' \
+ 'open-science-for-sim/main/src/notebooks/01_foss_sim/data/ed_arrivals.csv'
# visualise
ax = pd.read_csv(NSPP_PATH).plot(y='arrival_rate', x='period', rot=45,
kind='bar',figsize=(12,5), legend=False)
ax.set_xlabel('hour of day')
ax.set_ylabel('mean arrivals');
2.3 Resource counts#
Inter count variables representing the number of resources at each activity in the processes.
DEFAULT_N_TRIAGE = 1
DEFAULT_N_REG = 1
DEFAULT_N_EXAM = 3
DEFAULT_N_TRAUMA = 2
# Non-trauma cubicles
DEFAULT_N_CUBICLES_1 = 1
# trauma pathway cubicles
DEFAULT_N_CUBICLES_2 = 1
2.4 Simulation model run settings#
# default random number SET
DEFAULT_RNG_SET = None
N_STREAMS = 20
# default results collection period
DEFAULT_RESULTS_COLLECTION_PERIOD = 60 * 19
# number of replications.
DEFAULT_N_REPS = 5
# Show the a trace of simulated events
# not recommended when running multiple replications
DEFAULT_TRACE_LEVEL = 0
## deprecated...
TRACE = True
3. Trace functionality#
Rather than using the print built in function to output a simulated trace it is desirable to control the if trace messages are shown or not shown. The SimulatedTrace class is partly based on R’s simmer functionality for output. A user sets a trace_level threshold when creating an instance of the class. The class is callable like a function and replaces the print built in e.g
trace = SimulatedTrace()
trace("hello world")
It is then trivial to suppress messages to a user
trace = SimulatedTrace(trace_level=2)
# this will not be printed as log_level is 0 by default
trace("hello world")
# this will NOT be printed in this experiment as log_level is less than 2
trace("level 1 message", trace_level=1)
# this will be printed!
trace("level 2 message", trace_level=2)
class SimulatedTrace:
'''
Utility class for printing a trace as the
simulation model executes.
'''
def __init__(self, trace_level=0):
'''Simulated Trace
Log events as they happen.
Params:
-------
log_level: int, optional (default=0)
Minimum log level of a print statement
in order for it to be logged.
'''
self.trace_level = trace_level
def __call__(self, msg, trace_level=0):
'''Override callable.
This makes objects behave like functions.
decorates the print function. conditional
logic to print output or not.
Params:
------
msg: str
string to print to screen.
trace_level: int, optional (default=0)
minimum trace level in order for the message
to display
'''
if (trace_level >= self.trace_level):
print(msg)
4. Distribution classes#
To help with controlling sampling numpy distributions are packaged up into classes that allow easy control of random numbers.
Distributions included:
Exponential
Log Normal
Bernoulli
Normal
Uniform
class Exponential:
'''
Convenience class for the exponential distribution.
packages up distribution parameters, seed and random generator.
'''
def __init__(self, mean, random_seed=None):
'''
Constructor
Params:
------
mean: float
The mean of the exponential distribution
random_seed: int, optional (default=None)
A random seed to reproduce samples. If set to none then a unique
sample is created.
'''
self.rng = np.random.default_rng(seed=random_seed)
self.mean = mean
def sample(self, size=None):
'''
Generate a sample from the exponential distribution
Params:
-------
size: int, optional (default=None)
the number of samples to return. If size=None then a single
sample is returned.
'''
return self.rng.exponential(self.mean, size=size)
class Bernoulli:
'''
Convenience class for the Bernoulli distribution.
packages up distribution parameters, seed and random generator.
'''
def __init__(self, p, random_seed=None):
'''
Constructor
Params:
------
p: float
probability of drawing a 1
random_seed: int, optional (default=None)
A random seed to reproduce samples. If set to none then a unique
sample is created.
'''
self.rng = np.random.default_rng(seed=random_seed)
self.p = p
def sample(self, size=None):
'''
Generate a sample from the exponential distribution
Params:
-------
size: int, optional (default=None)
the number of samples to return. If size=None then a single
sample is returned.
'''
return self.rng.binomial(n=1, p=self.p, size=size)
class Lognormal:
"""
Encapsulates a lognormal distirbution
"""
def __init__(self, mean, stdev, random_seed=None):
"""
Params:
-------
mean: float
mean of the lognormal distribution
stdev: float
standard dev of the lognormal distribution
random_seed: int, optional (default=None)
Random seed to control sampling
"""
self.rng = np.random.default_rng(seed=random_seed)
mu, sigma = self.normal_moments_from_lognormal(mean, stdev**2)
self.mu = mu
self.sigma = sigma
def normal_moments_from_lognormal(self, m, v):
'''
Returns mu and sigma of normal distribution
underlying a lognormal with mean m and variance v
source: https://blogs.sas.com/content/iml/2014/06/04/simulate-lognormal
-data-with-specified-mean-and-variance.html
Params:
-------
m: float
mean of lognormal distribution
v: float
variance of lognormal distribution
Returns:
-------
(float, float)
'''
phi = math.sqrt(v + m**2)
mu = math.log(m**2/phi)
sigma = math.sqrt(math.log(phi**2/m**2))
return mu, sigma
def sample(self):
"""
Sample from the normal distribution
"""
return self.rng.lognormal(self.mu, self.sigma)
class Normal:
'''
Convenience class for the normal distribution.
packages up distribution parameters, seed and random generator.
Use the minimum parameter to truncate the distribution
'''
def __init__(self, mean, sigma, minimum=None, random_seed=None):
'''
Constructor
Params:
------
mean: float
The mean of the normal distribution
sigma: float
The stdev of the normal distribution
minimum: float, optional (default=None)
Used to truncate the distribution (e.g. to 0.0 or 0.5)
random_seed: int, optional (default=None)
A random seed to reproduce samples. If set to none then a unique
sample is created.
'''
self.rng = np.random.default_rng(seed=random_seed)
self.mean = mean
self.sigma = sigma
self.minimum = minimum
def sample(self, size=None):
'''
Generate a sample from the normal distribution
Params:
-------
size: int, optional (default=None)
the number of samples to return. If size=None then a single
sample is returned.
'''
samples = self.rng.normal(self.mean, self.sigma, size=size)
if self.minimum is None:
return samples
elif size is None:
return max(self.minimum, samples)
else:
# index of samples with negative value
neg_idx = np.where(samples < 0)[0]
samples[neg_idx] = self.minimum
return samples
class Uniform():
'''
Convenience class for the Uniform distribution.
packages up distribution parameters, seed and random generator.
'''
def __init__(self, low, high, random_seed=None):
'''
Constructor
Params:
------
low: float
lower range of the uniform
high: float
upper range of the uniform
random_seed: int, optional (default=None)
A random seed to reproduce samples. If set to none then a unique
sample is created.
'''
self.rand = np.random.default_rng(seed=random_seed)
self.low = low
self.high = high
def sample(self, size=None):
'''
Generate a sample from the uniform distribution
Params:
-------
size: int, optional (default=None)
the number of samples to return. If size=None then a single
sample is returned.
'''
return self.rand.uniform(low=self.low, high=self.high, size=size)
5. Model parameterisation#
For convienience a container class is used to hold the large number of model parameters. The Scenario class includes defaults these can easily be changed and at runtime to experiments with different designs.
class Scenario:
'''
Container class for scenario parameters/arguments
Passed to a model and its process classes
'''
def __init__(self, random_number_set=DEFAULT_RNG_SET,
n_triage=DEFAULT_N_TRIAGE,
n_reg=DEFAULT_N_REG,
n_exam=DEFAULT_N_EXAM,
n_trauma=DEFAULT_N_TRAUMA,
n_cubicles_1=DEFAULT_N_CUBICLES_1,
n_cubicles_2=DEFAULT_N_CUBICLES_2,
triage_mean=DEFAULT_TRIAGE_MEAN,
reg_mean=DEFAULT_REG_MEAN,
reg_var=DEFAULT_REG_VAR,
exam_mean=DEFAULT_EXAM_MEAN,
exam_var=DEFAULT_EXAM_VAR,
exam_min=DEFAULT_EXAM_MIN,
trauma_mean=DEFAULT_TRAUMA_MEAN,
trauma_treat_mean=DEFAULT_TRAUMA_TREAT_MEAN,
trauma_treat_var=DEFAULT_TRAUMA_TREAT_VAR,
non_trauma_treat_mean=DEFAULT_NON_TRAUMA_TREAT_MEAN,
non_trauma_treat_var=DEFAULT_NON_TRAUMA_TREAT_VAR,
non_trauma_treat_p=DEFAULT_NON_TRAUMA_TREAT_P,
prob_trauma=DEFAULT_PROB_TRAUMA,
trace_level=DEFAULT_TRACE_LEVEL):
'''
Create a scenario to parameterise the simulation model
Parameters:
-----------
random_number_set: int, optional (default=DEFAULT_RNG_SET)
Set to control the initial seeds of each stream of pseudo
random numbers used in the model.
n_triage: int
The number of triage cubicles
n_reg: int
The number of registration clerks
n_exam: int
The number of examination rooms
n_trauma: int
The number of trauma bays for stabilisation
n_cubicles_1: int
The number of non-trauma treatment cubicles
n_cubicles_2: int
The number of trauma treatment cubicles
triage_mean: float
Mean duration of the triage distribution (Exponential)
reg_mean: float
Mean duration of the registration distribution (Lognormal)
reg_var: float
Variance of the registration distribution (Lognormal)
exam_mean: float
Mean of the examination distribution (Normal)
exam_var: float
Variance of the examination distribution (Normal)
exam_min: float
The minimum value that an examination can take (Truncated Normal)
trauma_mean: float
Mean of the trauma stabilisation distribution (Exponential)
trauma_treat_mean: float
Mean of the trauma cubicle treatment distribution (Lognormal)
trauma_treat_var: float
Variance of the trauma cubicle treatment distribution (Lognormal)
non_trauma_treat_mean: float
Mean of the non trauma treatment distribution
non_trauma_treat_var: float
Variance of the non trauma treatment distribution
non_trauma_treat_p: float
Probability non trauma patient requires treatment
prob_trauma: float
probability that a new arrival is a trauma patient.
trace_level: int
minimum log level of the experiment. Controls
display of simulated trace.
'''
# sampling
self.random_number_set = random_number_set
# store parameters for sampling
self.triage_mean = triage_mean
self.reg_mean = reg_mean
self.reg_var = reg_var
self.exam_mean= exam_mean
self.exam_var = exam_var
self.exam_min = exam_min
self.trauma_mean = trauma_mean
self.trauma_treat_mean = trauma_treat_mean
self.trauma_treat_var = trauma_treat_var
self.non_trauma_treat_mean = non_trauma_treat_mean
self.non_trauma_treat_var = non_trauma_treat_var
self.non_trauma_treat_p = non_trauma_treat_p
self.prob_trauma = prob_trauma
self.trace = SimulatedTrace(trace_level)
self.init_sampling()
# count of each type of resource
self.init_resourse_counts(n_triage, n_reg, n_exam, n_trauma,
n_cubicles_1, n_cubicles_2)
def set_random_no_set(self, random_number_set):
'''
Controls the random sampling
Parameters:
----------
random_number_set: int
Used to control the set of pseudo random numbers
used by the distributions in the simulation.
'''
self.random_number_set = random_number_set
self.init_sampling()
def init_resourse_counts(self, n_triage, n_reg, n_exam, n_trauma,
n_cubicles_1, n_cubicles_2):
'''
Init the counts of resources to default values...
'''
self.n_triage = n_triage
self.n_reg = n_reg
self.n_exam = n_exam
self.n_trauma = n_trauma
# non-trauma (1), trauma (2) treatment cubicles
self.n_cubicles_1 = n_cubicles_1
self.n_cubicles_2 = n_cubicles_2
def init_sampling(self):
'''
Create the distributions used by the model and initialise
the random seeds of each.
'''
# MODIFICATION. Better method for producing n non-overlapping streams
seed_sequence = np.random.SeedSequence(self.random_number_set)
# Generate n high quality child seeds
self.seeds = seed_sequence.spawn(N_STREAMS)
# create distributions
# Triage duration
self.triage_dist = Exponential(self.triage_mean,
random_seed=self.seeds[0])
# Registration duration (non-trauma only)
self.reg_dist = Lognormal(self.reg_mean,
np.sqrt(self.reg_var),
random_seed=self.seeds[1])
# Evaluation (non-trauma only)
self.exam_dist = Normal(self.exam_mean,
np.sqrt(self.exam_var),
minimum=self.exam_min,
random_seed=self.seeds[2])
# Trauma/stablisation duration (trauma only)
self.trauma_dist = Exponential(self.trauma_mean,
random_seed=self.seeds[3])
# Non-trauma treatment
self.nt_treat_dist = Lognormal(self.non_trauma_treat_mean,
np.sqrt(self.non_trauma_treat_var),
random_seed=self.seeds[4])
# treatment of trauma patients
self.treat_dist = Lognormal(self.trauma_treat_mean,
np.sqrt(self.trauma_treat_var),
random_seed=self.seeds[5])
# probability of non-trauma patient requiring treatment
self.nt_p_treat_dist = Bernoulli(self.non_trauma_treat_p,
random_seed=self.seeds[6])
# probability of non-trauma versus trauma patient
self.p_trauma_dist = Bernoulli(self.prob_trauma,
random_seed=self.seeds[7])
# init sampling for non-stationary poisson process
self.init_nspp()
def init_nspp(self):
# read arrival profile
self.arrivals = pd.read_csv(NSPP_PATH)
self.arrivals['mean_iat'] = 60 / self.arrivals['arrival_rate']
# maximum arrival rate (smallest time between arrivals)
self.lambda_max = self.arrivals['arrival_rate'].max()
# thinning exponential
self.arrival_dist = Exponential(60.0 / self.lambda_max,
random_seed=self.seeds[8])
# thinning uniform rng
self.thinning_rng = Uniform(low=0.0, high=1.0,
random_seed=self.seeds[9])
6. Patient Pathways Process Logic#
simpy uses a process based worldview. We can easily create whatever logic - simple or complex for the model. Here the process logic for trauma and non-trauma patients is seperated into two classes TraumaPathway and NonTraumaPathway.
class TraumaPathway:
'''
Encapsulates the process a patient with severe injuries or illness.
These patients are signed into the ED and triaged as having severe injuries
or illness.
Patients are stabilised in resus (trauma) and then sent to Treatment.
Following treatment they are discharged.
'''
def __init__(self, identifier, env, args):
'''
Constructor method
Params:
-----
identifier: int
a numeric identifier for the patient.
env: simpy.Environment
the simulation environment
args: Scenario
Container class for the simulation parameters
'''
self.identifier = identifier
self.env = env
self.args = args
self.trace = args.trace
# metrics
self.arrival = -np.inf
self.wait_triage = -np.inf
self.wait_trauma = -np.inf
self.wait_treat = -np.inf
self.total_time = -np.inf
self.triage_duration = -np.inf
self.trauma_duration = -np.inf
self.treat_duration = -np.inf
def execute(self):
'''
simulates the major treatment process for a patient
1. request and wait for sign-in/triage
2. trauma
3. treatment
'''
# record the time of arrival and entered the triage queue
self.arrival = self.env.now
# request sign-in/triage
with self.args.triage.request() as req:
yield req
# record the waiting time for triage
self.wait_triage = self.env.now - self.arrival
self.trace(f'patient {self.identifier} triaged to trauma '
f'{self.env.now:.3f}')
# sample triage duration.
self.triage_duration = self.args.triage_dist.sample()
yield self.env.timeout(self.triage_duration)
self.triage_complete()
# record the time that entered the trauma queue
start_wait = self.env.now
# request trauma room
with self.args.trauma.request() as req:
yield req
# record the waiting time for trauma
self.wait_trauma = self.env.now - start_wait
# sample stablisation duration.
self.trauma_duration = self.args.trauma_dist.sample()
yield self.env.timeout(self.trauma_duration)
self.trauma_complete()
# record the time that entered the treatment queue
start_wait = self.env.now
# request treatment cubicle
with self.args.cubicle_2.request() as req:
yield req
# record the waiting time for trauma
self.wait_treat = self.env.now - start_wait
self.trace(f'treatment of patient {self.identifier} at '
f'{self.env.now:.3f}')
# sample treatment duration.
self.treat_duration = self.args.treat_dist.sample()
yield self.env.timeout(self.treat_duration)
self.treatment_complete()
# total time in system
self.total_time = self.env.now - self.arrival
def triage_complete(self):
'''
Triage complete event
'''
self.trace(f'triage {self.identifier} complete {self.env.now:.3f}; '
f'waiting time was {self.wait_triage:.3f}')
def trauma_complete(self):
'''
Patient stay in trauma is complete.
'''
self.trace(f'stabilisation of patient {self.identifier} at '
f'{self.env.now:.3f}')
def treatment_complete(self):
'''
Treatment complete event
'''
self.trace(f'patient {self.identifier} treatment complete {self.env.now:.3f}; '
f'waiting time was {self.wait_treat:.3f}')
class NonTraumaPathway:
'''
Encapsulates the process a patient with minor injuries and illness.
These patients are signed into the ED and triaged as having minor
complaints and streamed to registration and then examination.
Post examination 40% are discharged while 60% proceed to treatment.
Following treatment they are discharged.
'''
def __init__(self, identifier, env, args):
'''
Constructor method
Params:
-----
identifier: int
a numeric identifier for the patient.
env: simpy.Environment
the simulation environment
args: Scenario
Container class for the simulation parameters
'''
self.identifier = identifier
self.env = env
self.args = args
self.trace = args.trace
# triage resource
self.triage = args.triage
# metrics
self.arrival = -np.inf
self.wait_triage = -np.inf
self.wait_reg = -np.inf
self.wait_exam = -np.inf
self.wait_treat = -np.inf
self.total_time = -np.inf
self.triage_duration = -np.inf
self.reg_duration = -np.inf
self.exam_duration = -np.inf
self.treat_duration = -np.inf
def execute(self):
'''
simulates the non-trauma/minor treatment process for a patient
1. request and wait for sign-in/triage
2. patient registration
3. examination
4.1 40% discharged
4.2 60% treatment then discharge
'''
# record the time of arrival and entered the triage queue
self.arrival = self.env.now
# request sign-in/triage
with self.triage.request() as req:
yield req
# record the waiting time for triage
self.wait_triage = self.env.now - self.arrival
self.trace(f'patient {self.identifier} triaged to minors '
f'{self.env.now:.3f}')
# sample triage duration.
self.triage_duration = self.args.triage_dist.sample()
yield self.env.timeout(self.triage_duration)
self.trace(f'triage {self.identifier} complete {self.env.now:.3f}; '
f'waiting time was {self.wait_triage:.3f}')
# record the time that entered the registration queue
start_wait = self.env.now
# request registration clert
with self.args.registration.request() as req:
yield req
# record the waiting time for registration
self.wait_reg = self.env.now - start_wait
self.trace(f'registration of patient {self.identifier} at '
f'{self.env.now:.3f}')
# sample registration duration.
self.reg_duration = self.args.reg_dist.sample()
yield self.env.timeout(self.reg_duration)
self.trace(f'patient {self.identifier} registered at'
f'{self.env.now:.3f}; '
f'waiting time was {self.wait_reg:.3f}')
# record the time that entered the evaluation queue
start_wait = self.env.now
# request examination resource
with self.args.exam.request() as req:
yield req
# record the waiting time for registration
self.wait_exam = self.env.now - start_wait
self.trace(f'examination of patient {self.identifier} begins '
f'{self.env.now:.3f}')
# sample examination duration.
self.exam_duration = self.args.exam_dist.sample()
yield self.env.timeout(self.exam_duration)
self.trace(f'patient {self.identifier} examination complete '
f'at {self.env.now:.3f};'
f'waiting time was {self.wait_exam:.3f}')
# sample if patient requires treatment?
self.require_treat = self.args.nt_p_treat_dist.sample()
if self.require_treat:
# record the time that entered the treatment queue
start_wait = self.env.now
# request treatment cubicle
with self.args.cubicle_1.request() as req:
yield req
# record the waiting time for treatment
self.wait_treat = self.env.now - start_wait
self.trace(f'treatment of patient {self.identifier} begins '
f'{self.env.now:.3f}')
# sample treatment duration.
self.treat_duration = self.args.nt_treat_dist.sample()
yield self.env.timeout(self.treat_duration)
self.trace(f'patient {self.identifier} treatment complete '
f'at {self.env.now:.3f};'
f'waiting time was {self.wait_treat:.3f}')
# total time in system
self.total_time = self.env.now - self.arrival
7. Main model class#
The main class that a user interacts with to run the model is TreatmentCentreModel. This implements a .run() method, contains a simple algorithm for the non-stationary poission process for patients arrivals and inits instances of TraumaPathway or NonTraumaPathway depending on the arrival type.
class TreatmentCentreModel:
'''
The treatment centre model
Patients arrive at random to a treatment centre, are triaged
and then processed in either a trauma or non-trauma pathway.
'''
def __init__(self, args):
self.env = simpy.Environment()
self.args = args
self.trace = args.trace
self.init_resources()
self.patients = []
self.trauma_patients = []
self.non_trauma_patients = []
self.rc_period = None
self.results = None
def init_resources(self):
'''
Init the number of resources
and store in the arguments container object
Resource list:
1. Sign-in/triage bays
2. registration clerks
3. examination bays
4. trauma bays
5. non-trauma cubicles (1)
6. trauma cubicles (2)
'''
# sign/in triage
self.args.triage = simpy.Resource(self.env,
capacity=self.args.n_triage)
# registration
self.args.registration = simpy.Resource(self.env,
capacity=self.args.n_reg)
# examination
self.args.exam = simpy.Resource(self.env,
capacity=self.args.n_exam)
# trauma
self.args.trauma = simpy.Resource(self.env,
capacity=self.args.n_trauma)
# non-trauma treatment
self.args.cubicle_1 = simpy.Resource(self.env,
capacity=self.args.n_cubicles_1)
# trauma treatment
self.args.cubicle_2 = simpy.Resource(self.env,
capacity=self.args.n_cubicles_2)
def run(self, results_collection_period=DEFAULT_RESULTS_COLLECTION_PERIOD):
'''
Conduct a single run of the model in its current
configuration
Parameters:
----------
results_collection_period, float, optional
default = DEFAULT_RESULTS_COLLECTION_PERIOD
warm_up, float, optional (default=0)
length of initial transient period to truncate
from results.
Returns:
--------
None
'''
# setup the arrival generator process
self.env.process(self.arrivals_generator())
# store rc perio
self.rc_period = results_collection_period
# run
self.env.run(until=results_collection_period)
def arrivals_generator(self):
'''
Simulate the arrival of patients to the model
Patients either follow a TraumaPathway or
NonTraumaPathway simpy process.
Non stationary arrivals implemented via Thinning acceptance-rejection
algorithm.
'''
for patient_count in itertools.count():
# this give us the index of dataframe to use
t = int(self.env.now // 60) % self.args.arrivals.shape[0]
lambda_t = self.args.arrivals['arrival_rate'].iloc[t]
#set to a large number so that at least 1 sample taken!
u = np.Inf
interarrival_time = 0.0
# reject samples if u >= lambda_t / lambda_max
while u >= (lambda_t / self.args.lambda_max):
interarrival_time += self.args.arrival_dist.sample()
u = self.args.thinning_rng.sample()
# iat
yield self.env.timeout(interarrival_time)
self.trace(f'patient {patient_count} arrives at: {self.env.now:.3f}')
# sample if the patient is trauma or non-trauma
trauma = self.args.p_trauma_dist.sample()
if trauma:
# create and store a trauma patient to update KPIs.
new_patient = TraumaPathway(patient_count, self.env, self.args)
self.trauma_patients.append(new_patient)
else:
# create and store a non-trauma patient to update KPIs.
new_patient = NonTraumaPathway(patient_count, self.env,
self.args)
self.non_trauma_patients.append(new_patient)
# start the pathway process for the patient
self.env.process(new_patient.execute())
8. Logic to process end of run results.#
the class SimulationSummary accepts a TraumaCentreModel. At the end of a run it can be used calculate mean queuing times and the percentage of the total run that a resource was in use.
class SimulationSummary:
'''
End of run result processing logic of the simulation model
'''
def __init__(self, model):
'''
Constructor
Params:
------
model: TraumaCentreModel
The model.
'''
self.model = model
self.args = model.args
self.results = None
def process_run_results(self):
'''
Calculates statistics at end of run.
'''
self.results = {}
# list of all patients
patients = self.model.non_trauma_patients + self.model.trauma_patients
# mean triage times (both types of patient)
mean_triage_wait = self.get_mean_metric('wait_triage', patients)
# triage utilisation (both types of patient)
triage_util = self.get_resource_util('triage_duration',
self.args.n_triage,
patients)
# mean waiting time for registration (non_trauma)
mean_reg_wait = self.get_mean_metric('wait_reg',
self.model.non_trauma_patients)
# registration utilisation (trauma)
reg_util = self.get_resource_util('reg_duration',
self.args.n_reg,
self.model.non_trauma_patients)
# mean waiting time for examination (non_trauma)
mean_wait_exam = self.get_mean_metric('wait_exam',
self.model.non_trauma_patients)
# examination utilisation (non-trauma)
exam_util = self.get_resource_util('exam_duration',
self.args.n_exam,
self.model.non_trauma_patients)
# mean waiting time for treatment (non-trauma)
mean_treat_wait = self.get_mean_metric('wait_treat',
self.model.non_trauma_patients)
# treatment utilisation (non_trauma)
treat_util1 = self.get_resource_util('treat_duration',
self.args.n_cubicles_1,
self.model.non_trauma_patients)
# mean total time (non_trauma)
mean_total = self.get_mean_metric('total_time',
self.model.non_trauma_patients)
# mean waiting time for trauma
mean_trauma_wait = self.get_mean_metric('wait_trauma',
self.model.trauma_patients)
# trauma utilisation (trauma)
trauma_util = self.get_resource_util('trauma_duration',
self.args.n_trauma,
self.model.trauma_patients)
# mean waiting time for treatment (rauma)
mean_treat_wait2 = self.get_mean_metric('wait_treat',
self.model.trauma_patients)
# treatment utilisation (trauma)
treat_util2 = self.get_resource_util('treat_duration',
self.args.n_cubicles_2,
self.model.trauma_patients)
# mean total time (trauma)
mean_total2 = self.get_mean_metric('total_time',
self.model.trauma_patients)
self.results = {'00_arrivals':len(patients),
'01a_triage_wait': mean_triage_wait,
'01b_triage_util': triage_util,
'02a_registration_wait':mean_reg_wait,
'02b_registration_util': reg_util,
'03a_examination_wait':mean_wait_exam,
'03b_examination_util': exam_util,
'04a_treatment_wait(non_trauma)':mean_treat_wait,
'04b_treatment_util(non_trauma)':treat_util1,
'05_total_time(non-trauma)':mean_total,
'06a_trauma_wait':mean_trauma_wait,
'06b_trauma_util':trauma_util,
'07a_treatment_wait(trauma)':mean_treat_wait2,
'07b_treatment_util(trauma)':treat_util2,
'08_total_time(trauma)':mean_total2,
'09_throughput': self.get_throughput(patients)}
def get_mean_metric(self, metric, patients):
'''
Calculate mean of the performance measure for the
select cohort of patients,
Only calculates metrics for patients where it has been
measured.
Params:
-------
metric: str
The name of the metric e.g. 'wait_treat'
patients: list
A list of patients
'''
mean = np.array([getattr(p, metric) for p in patients
if getattr(p, metric) > -np.inf]).mean()
return mean
def get_resource_util(self, metric, n_resources, patients):
'''
Calculate proportion of the results collection period
where a resource was in use.
Done by tracking the duration by patient.
Only calculates metrics for patients where it has been
measured.
Params:
-------
metric: str
The name of the metric e.g. 'treatment_duration'
patients: list
A list of patients
'''
total = np.array([getattr(p, metric) for p in patients
if getattr(p, metric) > -np.inf]).sum()
return total / (self.model.rc_period * n_resources)
def get_throughput(self, patients):
'''
Returns the total number of patients that have successfully
been processed and discharged in the treatment centre
(they have a total time record)
Params:
-------
patients: list
list of all patient objects simulated.
Returns:
------
float
'''
return len([p for p in patients if p.total_time > -np.inf])
def summary_frame(self):
'''
Returns run results as a pandas.DataFrame
Returns:
-------
pd.DataFrame
'''
#append to results df
if self.results is None:
self.process_run_results()
df = pd.DataFrame({'1':self.results})
df = df.T
df.index.name = 'rep'
return df
9. Model execution#
We note that there are many ways to setup a simpy model and execute it (that is part of its fantastic flexibility). The organisation of code we show below is based on our experience of using the package in practice. The approach also allows for easy parallisation over multiple CPU cores using joblib.
We include two functions. single_run() and multiple_replications. The latter is used to repeatedly call and process the results from single_run.
def single_run(scenario, rc_period=DEFAULT_RESULTS_COLLECTION_PERIOD,
random_no_set=DEFAULT_RNG_SET):
'''
Perform a single run of the model and return the results
Parameters:
-----------
scenario: Scenario object
The scenario/paramaters to run
rc_period: int
The length of the simulation run that collects results
random_no_set: int or None, optional (default=DEFAULT_RNG_SET)
Controls the set of random seeds used by the stochastic parts of the
model. Set to different ints to get different results. Set to None
for a random set of seeds.
Returns:
--------
pandas.DataFrame:
results from single run.
'''
# set random number set - this controls sampling for the run.
scenario.set_random_no_set(random_no_set)
# create an instance of the model
model = TreatmentCentreModel(scenario)
# run the model
model.run(results_collection_period=rc_period)
# run results
summary = SimulationSummary(model)
summary_df = summary.summary_frame()
return summary_df
def multiple_replications(scenario, rc_period=DEFAULT_RESULTS_COLLECTION_PERIOD,
n_reps=5):
'''
Perform multiple replications of the model.
Params:
------
scenario: Scenario
Parameters/arguments to configurethe model
rc_period: float, optional (default=DEFAULT_RESULTS_COLLECTION_PERIOD)
results collection period.
the number of minutes to run the model to collect results
n_reps: int, optional (default=DEFAULT_N_REPS)
Number of independent replications to run.
Returns:
--------
pandas.DataFrame
'''
results = [single_run(scenario, rc_period, random_no_set=rep)
for rep in range(n_reps)]
#format and return results in a dataframe
df_results = pd.concat(results)
df_results.index = np.arange(1, len(df_results)+1)
df_results.index.name = 'rep'
return df_results
9.1 Single run of the model#
The script below performs a single replication of the simulation model.
Try:
Changing the
random_no_setof thesingle_runcall.Assigning the value
TruetoTRACE
# Change this to True to see a trace...
#TRACE = False
# create the default scenario.
args = Scenario(trace_level=2)
# use the single_run() func
# try changing `random_no_set` to see different run results
print('Running simulation ...', end=' => ')
results = single_run(args, random_no_set=42)
print('simulation complete.')
# show results (transpose replication for easier view)
results.T
Running simulation ... => simulation complete.
| rep | 1 |
|---|---|
| 00_arrivals | 209.000000 |
| 01a_triage_wait | 16.622674 |
| 01b_triage_util | 0.527512 |
| 02a_registration_wait | 111.161345 |
| 02b_registration_util | 0.801061 |
| 03a_examination_wait | 24.927965 |
| 03b_examination_util | 0.851285 |
| 04a_treatment_wait(non_trauma) | 172.435861 |
| 04b_treatment_util(non_trauma) | 0.845652 |
| 05_total_time(non-trauma) | 248.848441 |
| 06a_trauma_wait | 201.144403 |
| 06b_trauma_util | 0.919830 |
| 07a_treatment_wait(trauma) | 22.620880 |
| 07b_treatment_util(trauma) | 0.493576 |
| 08_total_time(trauma) | 310.384648 |
| 09_throughput | 155.000000 |
9.2 Multiple independent replications#
Given the set up it is now easy to perform multiple replications of the model.
%%time
args = Scenario(trace_level=2)
#run multiple replications.
#by default it runs 5 replications.
print('Running multiple replications', end=' => ')
results = multiple_replications(args, n_reps=50)
print('done.\n')
results.head(3)
Running multiple replications =>
done.
CPU times: user 1.32 s, sys: 60.9 ms, total: 1.38 s
Wall time: 4.83 s
| 00_arrivals | 01a_triage_wait | 01b_triage_util | 02a_registration_wait | 02b_registration_util | 03a_examination_wait | 03b_examination_util | 04a_treatment_wait(non_trauma) | 04b_treatment_util(non_trauma) | 05_total_time(non-trauma) | 06a_trauma_wait | 06b_trauma_util | 07a_treatment_wait(trauma) | 07b_treatment_util(trauma) | 08_total_time(trauma) | 09_throughput | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rep | ||||||||||||||||
| 1 | 230.0 | 24.280943 | 0.613250 | 103.242292 | 0.854504 | 31.089680 | 0.861719 | 152.483394 | 0.890904 | 234.759918 | 236.508444 | 1.028887 | 13.701783 | 0.607996 | 346.698079 | 171.0 |
| 2 | 227.0 | 57.120114 | 0.621348 | 90.002385 | 0.836685 | 14.688492 | 0.847295 | 120.245474 | 0.912127 | 233.882040 | 133.813901 | 0.834124 | 3.715446 | 0.367507 | 301.521195 | 161.0 |
| 3 | 229.0 | 28.659383 | 0.573698 | 112.242503 | 0.848514 | 21.374092 | 0.856306 | 94.019885 | 0.868888 | 208.361290 | 276.422566 | 0.874245 | 12.252175 | 0.464740 | 440.515502 | 167.0 |
# summarise the results (2.dp)
results.mean().round(2)
00_arrivals 227.72
01a_triage_wait 35.24
01b_triage_util 0.61
02a_registration_wait 105.57
02b_registration_util 0.84
03a_examination_wait 25.55
03b_examination_util 0.85
04a_treatment_wait(non_trauma) 136.66
04b_treatment_util(non_trauma) 0.87
05_total_time(non-trauma) 234.34
06a_trauma_wait 151.68
06b_trauma_util 0.83
07a_treatment_wait(trauma) 14.31
07b_treatment_util(trauma) 0.50
08_total_time(trauma) 292.28
09_throughput 162.16
dtype: float64
9.3 Visualise replications#
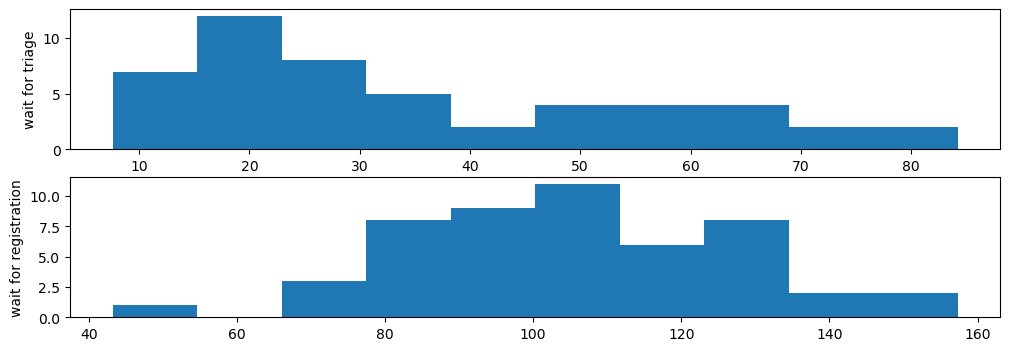
fig, ax = plt.subplots(2, 1, figsize=(12,4))
ax[0].hist(results['01a_triage_wait']);
ax[0].set_ylabel('wait for triage')
ax[1].hist(results['02a_registration_wait']);
ax[1].set_ylabel('wait for registration');
## 10. Scenario Analysis
The structured approach we took to organising our simpy model allows us to easily experiment with alternative scenarios. We could employ a formal experimental design if needed. For simplicity here we will limit ourselves by running user chosen competing scenarios and compare their mean performance to the base case.
Note that we have our
simpymodel includes an implementation of Common Random Numbers across scenarios.
def get_scenarios(trace_level=2):
'''
Creates a dictionary object containing
objects of type `Scenario` to run.
Returns:
--------
dict
Contains the scenarios for the model
'''
scenarios = {}
scenarios['base'] = Scenario(trace_level=trace_level)
# extra triage capacity
scenarios['triage+1'] = Scenario(n_triage=DEFAULT_N_TRIAGE+1,
trace_level=trace_level)
# extra examination capacity
scenarios['exam+1'] = Scenario(n_exam=DEFAULT_N_EXAM+1,
trace_level=trace_level)
# extra non-trauma treatment capacity
scenarios['treat+1'] = Scenario(n_cubicles_1=DEFAULT_N_CUBICLES_1+1,
trace_level=trace_level)
scenarios['triage+exam'] = Scenario(n_triage=DEFAULT_N_TRIAGE+1,
n_exam=DEFAULT_N_EXAM+1,
trace_level=trace_level)
return scenarios
def run_scenario_analysis(scenarios, rc_period, n_reps):
'''
Run each of the scenarios for a specified results
collection period and replications.
Params:
------
scenarios: dict
dictionary of Scenario objects
rc_period: float
model run length
n_rep: int
Number of replications
'''
print('Scenario Analysis')
print(f'No. Scenario: {len(scenarios)}')
print(f'Replications: {n_reps}')
scenario_results = {}
for sc_name, scenario in scenarios.items():
print(f'Running {sc_name}', end=' => ')
replications = multiple_replications(scenario, rc_period=rc_period,
n_reps=n_reps)
print('done.\n')
#save the results
scenario_results[sc_name] = replications
print('Scenario analysis complete.')
return scenario_results
10.1 Script to run scenario analysis#
#number of replications
N_REPS = 20
#get the scenarios
scenarios = get_scenarios()
#run the scenario analysis
scenario_results = run_scenario_analysis(scenarios,
DEFAULT_RESULTS_COLLECTION_PERIOD,
N_REPS)
Scenario Analysis
No. Scenario: 5
Replications: 20
Running base =>
done.
Running triage+1 =>
done.
Running exam+1 =>
done.
Running treat+1 =>
done.
Running triage+exam =>
done.
Scenario analysis complete.
def scenario_summary_frame(scenario_results):
'''
Mean results for each performance measure by scenario
Parameters:
----------
scenario_results: dict
dictionary of replications.
Key identifies the performance measure
Returns:
-------
pd.DataFrame
'''
columns = []
summary = pd.DataFrame()
for sc_name, replications in scenario_results.items():
summary = pd.concat([summary, replications.mean()], axis=1)
columns.append(sc_name)
summary.columns = columns
return summary
# as well as rounding you may want to rename the cols/rows to
# more readable alternatives.
summary_frame = scenario_summary_frame(scenario_results)
summary_frame.round(2)
| base | triage+1 | exam+1 | treat+1 | triage+exam | |
|---|---|---|---|---|---|
| 00_arrivals | 227.25 | 227.25 | 227.25 | 227.25 | 227.25 |
| 01a_triage_wait | 32.56 | 1.26 | 32.56 | 32.56 | 1.26 |
| 01b_triage_util | 0.61 | 0.31 | 0.61 | 0.61 | 0.31 |
| 02a_registration_wait | 104.69 | 131.82 | 104.69 | 104.69 | 131.82 |
| 02b_registration_util | 0.85 | 0.85 | 0.85 | 0.85 | 0.85 |
| 03a_examination_wait | 23.36 | 24.44 | 0.14 | 23.36 | 0.14 |
| 03b_examination_util | 0.86 | 0.86 | 0.67 | 0.86 | 0.67 |
| 04a_treatment_wait(non_trauma) | 130.73 | 133.09 | 144.50 | 2.15 | 147.81 |
| 04b_treatment_util(non_trauma) | 0.88 | 0.88 | 0.88 | 0.62 | 0.88 |
| 05_total_time(non-trauma) | 229.04 | 226.38 | 218.29 | 187.98 | 215.06 |
| 06a_trauma_wait | 166.98 | 189.67 | 166.98 | 166.98 | 189.67 |
| 06b_trauma_util | 0.84 | 0.86 | 0.84 | 0.84 | 0.86 |
| 07a_treatment_wait(trauma) | 14.39 | 14.77 | 14.39 | 14.39 | 14.77 |
| 07b_treatment_util(trauma) | 0.52 | 0.52 | 0.52 | 0.52 | 0.52 |
| 08_total_time(trauma) | 306.46 | 298.22 | 306.46 | 306.46 | 298.22 |
| 09_throughput | 165.85 | 166.65 | 169.10 | 196.85 | 169.85 |
11. Script to produce formatted LaTeX table for paper#
HEADER_URL = 'data/tbl_row_headers.csv'
WAIT = 'wait'
# filter for waiting times only and round to 2dp
waiting_times = summary_frame[summary_frame.index.str.contains(WAIT)].round(2)
# load formatted table headers
row_headers = pd.read_csv(HEADER_URL)
# merge and format headers
waiting_times = waiting_times.reset_index()
waiting_times = pd.concat([row_headers, waiting_times], axis=1)
waiting_times = waiting_times.set_index(row_headers.columns[0])
waiting_times = waiting_times.drop(['index'], axis=1)
waiting_times = waiting_times.reset_index()
waiting_times
| Mean waiting time (mins) | base | triage+1 | exam+1 | treat+1 | triage+exam | |
|---|---|---|---|---|---|---|
| 0 | Triage | 32.56 | 1.26 | 32.56 | 32.56 | 1.26 |
| 1 | Registation | 104.69 | 131.82 | 104.69 | 104.69 | 131.82 |
| 2 | Examination | 23.36 | 24.44 | 0.14 | 23.36 | 0.14 |
| 3 | Non-trauma treatment | 130.73 | 133.09 | 144.50 | 2.15 | 147.81 |
| 4 | Trauma stabilisation | 166.98 | 189.67 | 166.98 | 166.98 | 189.67 |
| 5 | Trauma treatment | 14.39 | 14.77 | 14.39 | 14.39 | 14.77 |
# output to file as LaTeX
OUTPUT_FILE = 'output/table_3.txt'
LABEL = 'tab:table3'
CAPTION = 'Simulation results that can be verified by our example reproducible pipeline.'
waiting_times.to_latex(OUTPUT_FILE, index=False, label=LABEL, caption=CAPTION)
# modify LaTeX file -> convert \caption to \tbl
with fileinput.FileInput(OUTPUT_FILE, inplace=1) as latex_file:
for line in latex_file:
line = line.replace('caption', 'tbl')
print(line, end='')
# view LaTeX to verify
with open(OUTPUT_FILE, "r+") as file1:
print(file1.read())
\begin{table}
\tbl{Simulation results that can be verified by our example reproducible pipeline.}
\label{tab:table3}
\begin{tabular}{lrrrrr}
\toprule
Mean waiting time (mins) & base & triage+1 & exam+1 & treat+1 & triage+exam \\
\midrule
Triage & 32.560000 & 1.260000 & 32.560000 & 32.560000 & 1.260000 \\
Registation & 104.690000 & 131.820000 & 104.690000 & 104.690000 & 131.820000 \\
Examination & 23.360000 & 24.440000 & 0.140000 & 23.360000 & 0.140000 \\
Non-trauma treatment & 130.730000 & 133.090000 & 144.500000 & 2.150000 & 147.810000 \\
Trauma stabilisation & 166.980000 & 189.670000 & 166.980000 & 166.980000 & 189.670000 \\
Trauma treatment & 14.390000 & 14.770000 & 14.390000 & 14.390000 & 14.770000 \\
\bottomrule
\end{tabular}
\end{table}
